Transmembrane helix prediction
Many proteins are integral membrane proteins. Most membrane proteins have hydrophobic regions which span the hydrophobic core of the membrane bi-layer and hydrophilic regions located on the outside or the inside of the membrane. Many receptor proteins have several transmembrane helices spanning the cellular membrane.
For prediction of transmembrane helices, CLC Genomics Workbench uses TMHMM version 2.0 [Krogh et al., 2001] located at http://www.cbs.dtu.dk/services/TMHMM/, thus an active internet connection is required to run the transmembrane helix prediction. Additional information on THMHH and Center for Biological Sequence analysis (CBS) can be found at http://www.cbs.dtu.dk and in the original research paper [Krogh et al., 2001].
In order to use the transmembrane helix prediction, you need to download the plug-in using the plug-in manager.
When the plug-in is downloaded and installed, you can use it to predict transmembrane helices:
Select a protein sequence | Toolbox in the Menu
Bar | Classical Sequence Analysis (![]() ) | Protein Analysis (
) | Protein Analysis (![]() )|
Transmembrane Helix Prediction (
)|
Transmembrane Helix Prediction (![]() )
)
or right-click a protein sequence | Toolbox |
Classical Sequence Analysis (![]() ) | Protein Analysis (
) | Protein Analysis (![]() )| Transmembrane
Helix Prediction (
)| Transmembrane
Helix Prediction (![]() )
)
If a sequence was selected before choosing the Toolbox action, this sequence is now listed in the Selected Elements window of the dialog. Use the arrows to add or remove sequences or sequence lists from the selected elements.
The predictions obtained can either be shown as annotations on the sequence, in a table or as the detailed and text output from the TMHMM method.
- Add annotations to sequence
- Create table
- Text
You can perform the analysis on several protein sequences at a time. This will add annotations to all the sequences and open a view for each sequence if a transmembrane helix is found. If a transmembrane helix is not found a dialog box will be presented.
After running the prediction as described above, the protein sequence will show predicted transmembrane helices as annotations on the original sequence (see figure 16.8). Moreover, annotations showing the topology will be shown. That is, which part the proteins is located on the inside or on the outside.
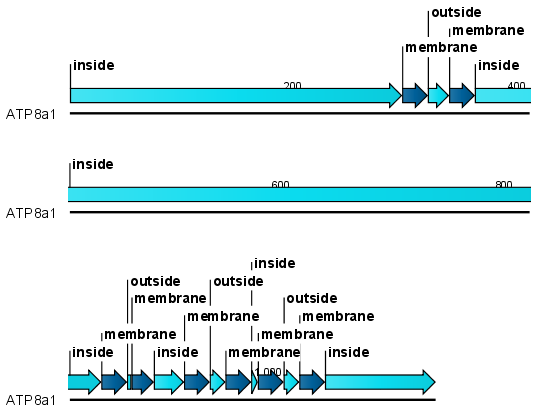
Figure 16.8: Transmembrane segments shown as annotation on the sequence and the topology.
Each annotation will carry a tooltip note saying that the
corresponding annotation is predicted with TMHMM version 2.0.
Additional notes can be added through the Edit
annotation (![]() ) right-click mouse menu.
Add and modify annotations.
) right-click mouse menu.
Add and modify annotations.
Undesired annotations can be removed through the Delete
Annotation (![]() ) right-click mouse menu.
Removing annotations.
) right-click mouse menu.
Removing annotations.
