Adding annotations
Adding annotations to a sequence
can be done in two ways:
open the sequence in a sequence view (double-click in
the Navigation Area) | make a selection covering the part of
the sequence you want to annotate10.1 | right-click the
selection | Add Annotation (![]() )
)
or select the sequence in the Navigation Area |
Show (![]() ) | Annotations (
) | Annotations (![]() ) | right click anywhere in the annotation table |
select New Annotation (
) | right click anywhere in the annotation table |
select New Annotation (![]() )
)
This will display a dialog like the one in figure 10.12.
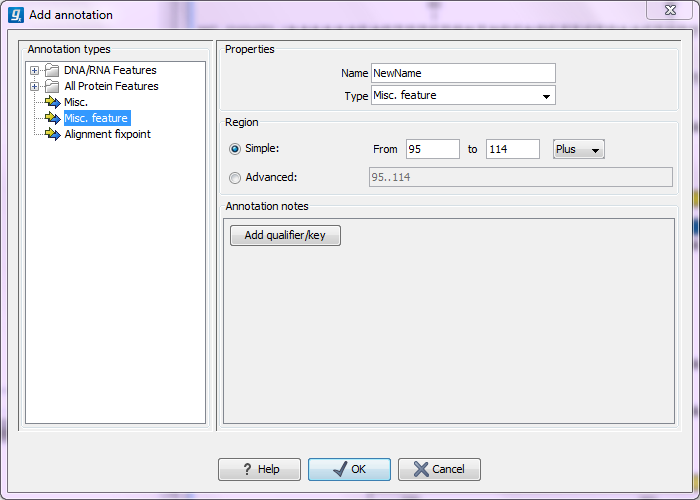
Figure 10.12: The Add Annotation dialog.
The left-hand part of the dialog lists a number of Annotation types. When you have selected an annotation type, it appears in Type to the right. You can also select an annotation directly in this list. Choosing an annotation type is mandatory. If you wish to use an annotation type which is not present in the list, simply enter this type into the Type field 10.2.
The right-hand part of the dialog contains the following text fields:
- Name. The name of the annotation which can be shown on the label in the sequence views. (Whether the name is actually shown depends on the Annotation Layout preferences, see Annotation Layout).
- Type. Reflects the left-hand part of the dialog as described above. You can also choose directly in this list or type your own annotation type.
- Region. If you have already made a selection,
this field will show the positions of the selection. You can modify
the region further using the conventions of DDBJ, EMBL and GenBank.
The following are examples of how to use the syntax (based on
http://www.ncbi.nlm.nih.gov/collab/FT/):
- 467. Points to a single residue in the presented sequence.
- 340..565. Points to a continuous range of residues bounded by and including the starting and ending residues.
- <345..500. Indicates that the exact lower boundary point of a region is unknown. The location begins at some residue previous to the first residue specified (which is not necessarily contained in the presented sequence) and continues up to and including the ending residue.
- <1..888. The region starts before the first sequenced residue and continues up to and including residue 888.
- 1..>888. The region starts at the first sequenced residue and continues beyond residue 888.
- (102.110). Indicates that the exact location is unknown, but that it is one of the residues between residues 102 and 110, inclusive.
- 123^124. Points to a site between residues 123 and 124.
- join(12..78,134..202). Regions 12 to 78 and 134 to 202 should be joined to form one contiguous sequence.
- complement(34..126) Start at the residue complementary to 126 and finish at the residue complementary to residue 34 (the region is on the strand complementary to the presented strand).
- complement(join(2691..4571,4918..5163)). Joins regions 2691 to 4571 and 4918 to 5163, then complements the joined segments (the region is on the strand complementary to the presented strand).
- join(complement(4918..5163),complement(2691..4571)). Complements regions 4918 to 5163 and 2691 to 4571, then joins the complemented segments (the region is on the strand complementary to the presented strand).
- Annotations. In this field, you can add more information about the annotation like comments and links. Click the Add qualifier/key button to enter information.
Select a qualifier which describes the kind of information you wish
to add. If an appropriate qualifier is not present in the list, you
can type your own qualifier. The pre-defined qualifiers are derived
from the GenBank format. You can add as many qualifier/key lines as
you wish by clicking the button. Redundant lines can be removed by
clicking the delete icon (
 ). The information entered on
these lines is shown in the annotation table (see View annotation
table) and in the yellow box which
appears when you place the mouse cursor on the annotation. If you
write a hyperlink in the Key text field, like e.g.
"www.clcbio.com", it will be recognized as a hyperlink. Clicking the
link in the annotation table will open a web browser.
). The information entered on
these lines is shown in the annotation table (see View annotation
table) and in the yellow box which
appears when you place the mouse cursor on the annotation. If you
write a hyperlink in the Key text field, like e.g.
"www.clcbio.com", it will be recognized as a hyperlink. Clicking the
link in the annotation table will open a web browser.
Click OK to add the annotation.
Note! The annotation will be included if you export the sequence in GenBank, Swiss-Prot or CLC format. When exporting in other formats, annotations are not preserved in the exported file.
Footnotes
- ... annotate10.1
- (See Selection on how to make selections that are not contiguous.)
- ... field10.2
- Note that your own annotation types will be converted to "unsure" when exporting in GenBank format. As long as you use the sequence in CLC format, you own annotation type will be preserved
