Add sequences to an existing contig
This section describes how to assemble sequences to an existing contig. This feature can be used for example to provide a steady work-flow when a number of exons from the same gene are sequenced one at a time and assembled to a reference sequence.
Note that the new sequences will be added to the existing contig which will not be extended. If the new sequences extend beyond the existing contig, they will be cut off.
To start the assembly:
select one contig and a number of sequences |
Toolbox in the Menu Bar | Molecular Biology Tools (![]() ) | Sequencing Data Analysis (
) | Sequencing Data Analysis (![]() )| Add Sequences to
Contig (
)| Add Sequences to
Contig (![]() )
)
or right-click in the empty white area of the contig |
Add Sequences to Contig (![]() )
)
This opens a dialog where you can alter your choice of sequences which you want to assemble. You can also add sequence lists.
Often, the results of the assembly will be better if the sequences are trimmed first (see Automatic trimming).
When the elements are selected, click Next, and you will see the dialog shown in figure 18.9
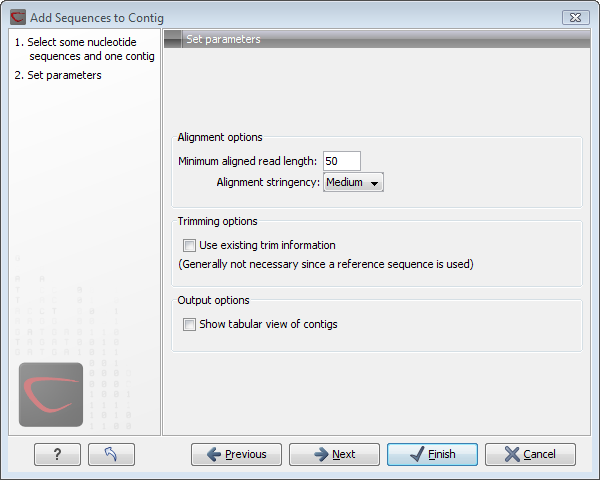
Figure 18.9: Setting assembly parameters when assembling to an existing contig.
The options in this dialog are similar to the options that are available when assembling to a reference sequence.
Click Next if you wish to adjust how to handle the results. If not, click Finish. This will start the assembly process.See View and edit contigs on how to use the resulting contig.
Note that the new sequences will be added to the existing contig which will not be extended. If the new sequences extend beyond the existing contig, they will be cut off.
