Gene fusion reporting
When using paired data, there is also an option to create a table summarizing the evidence for gene fusions. An example is shown in figure 27.11.
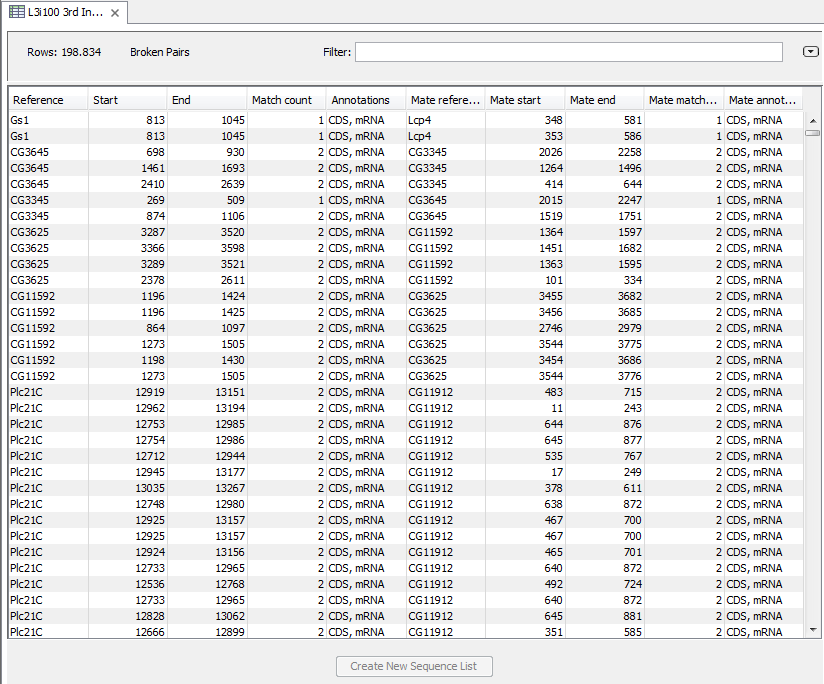
Figure 27.11: An example of a gene fusion table.
The table includes one row for each broken read pair where the reads are placed in different genes. The Minimum read count option is used to make sure that only combinations of genes supported by at least this number of read pairs are included. The default value is 5, which means that at least 5 broken pairs need to connect two genes, in order to include the broken pairs for this gene combination in the result table.
The result table shows the following information for each read:
- Reference. The name of the reference sequence (the name of the gene).
- Start. The position where the alignment of the read starts.
- End. The position where the alignment of the read ends.
- Match count. How many other positions this read could have mapped to equally well.
- Annotations. The type of annotation covering the read. This will show whether the read is in an exon or not.
