The Perform QIAseq RNA Fusion XP Analysis template workflows
The Perform QIAseq RNA Fusion XP Analysis (Illumina) or (Ion Torrent) template workflows can be found in the Toolbox at:
Template Workflows | Biomedical Workflows (![]() ) | QIAseq Sample Analysis (
) | QIAseq Sample Analysis (![]() ) | QIAseq Analysis Workflows (
) | QIAseq Analysis Workflows (![]() ) | Perform QIAseq RNA Fusion XP Analysis (Illumina/Ion Torrent) (
) | Perform QIAseq RNA Fusion XP Analysis (Illumina/Ion Torrent) (![]() )
)
The workflows are designed for use together with the QIAseq RNA Fusion XP panels (JHS-001Z, JHS-002Z, JHS-003Z, JHS-004Z, JHS-005Z, JHS-3001Z and JHS3002Z). The panels allow fusions detection, variant calls, and expression values to be generated from the same RNA sample, and therefore improve on the capabilities of the existing QIAseq RNAscan panels. These extended capabilities are supported by primers which have been annotated with the purpose for which they are intended, "Fusion", "Variant" and "GEX". Gene expresisons are calculated based on reads matching "GEX" anotated primers, fusions are detected using reads matching "Fusion" annotated primers while while SNV/IndDel variants are called based on all the reads.
The workflow requires the panel primers to be specified, either by selecting them from the Reference Data Set QIAseq RNA Fusion XP Panels hg38 for QIAGEN panels or, for custom panels, by importing them as described in Import QIAGEN Primers.
Double-click on the Perform QIAseq RNA Fusion XP Analysis template workflow to run the analysis.
If you are connected to a CLC Server via your Workbench, you will be asked where you would like to run the analysis. We recommend that you run the analysis on a CLC Server when possible.
In the Select RNA Reads dialog, specify the RNA reads that should be analyzed (figure 7.5).
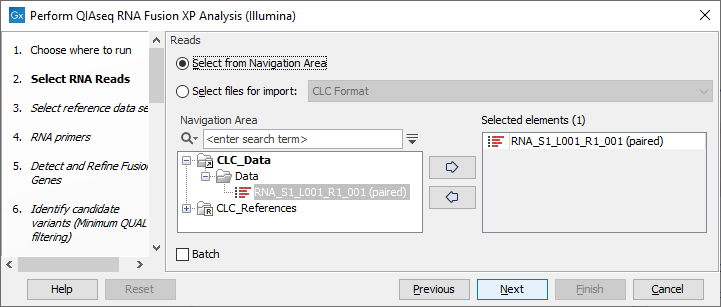
Figure 7.5: Select the sequencing reads by double-clicking on the file name or by clicking once on the file name and then on the arrow pointing to the right hand side.
The following dialog helps you set up the relevant Reference Data Set. If you have not downloaded the Reference Data Set yet, the dialog will suggest the relevant data set and offer the opportunity to download it using the Download to Workbench button. (figure 7.6).
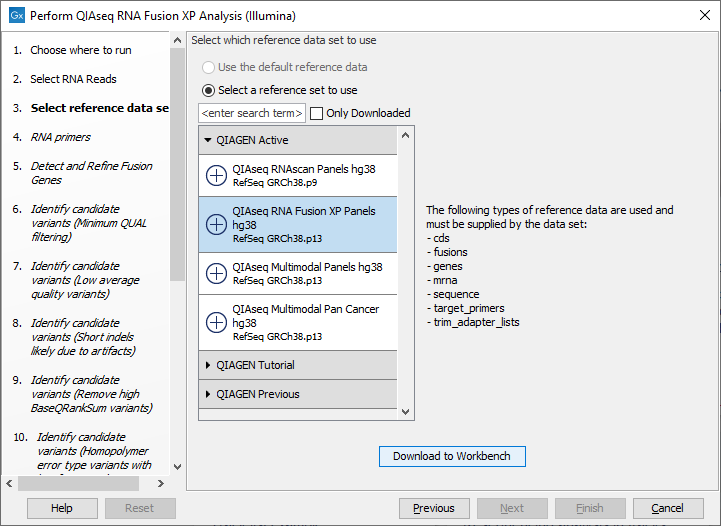
Figure 7.6: The relevant Reference Data Set is highlighted; in the text to the right, the types of reference needed by the workflow are listed.
Note that if you wish to Cancel or Resume the Download, you can close the template workflow and open the Reference Data Manager where the Cancel, Pause and Resume buttons are available.
If the Reference Data Set was previously downloaded, the option "Use the default reference data" is available and will ensure the relevant data set is used. You can always check the "Select a reference set to use" option to be able to specify another Reference Data Set than the one suggested.
In the RNA primers dialog, select primers that correspond to the panel that was used to generate the reads (figure 7.7).
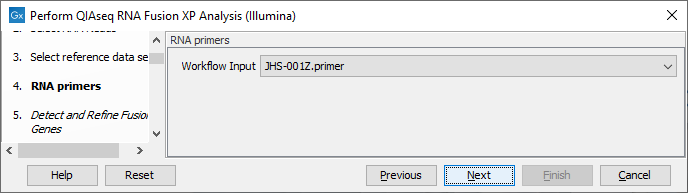
Figure 7.7: Select the primer track for the relevant panel.
In the Detect and Refine Fusion Genes dialog, it is possible to change the Promiscuity threshold, i.e., the maximum number of different fusion partners reported for a gene. You can also check for exon skippings by enabling the Detect exon skippings option, as well as check for fusions with novel exon boundaries by enabeling the Detect fusions with novel exon boundaries option.
The parameters for variant detection are not adjustable and have been set to generate an initial pool of all potential variants. These are then passed through a series of filters to remove variants that are suspected artifacts. Variants failing to meet the (adjustable) thresholds for quality, read direction bias, location (low frequency indels within homopolymer stretches), frequency or coverage would not be included in the filtered output.
Some filters only remove alternative alleles - and not reference alleles - as this potentially leads to wrong interpretation of variants by the VCF exporter where such variants could be misinterpreted as hemizygote when the reference allele is missing.
Finally, in the last wizard step, choose to Save the results of the workflow and specify a location in the Navigation Area before clicking Finish.
Subsections
