Create Tree from Comparison
Create Tree from Comparison builds a tree from a Pairwise Comparison such as those generated by Create Average Nucleotide Identity Comparison tool.
To run Create Tree from Comparison, go to:
Tools | Whole Genome Alignment (![]() ) | Create Tree from Comparison (
) | Create Tree from Comparison (![]() )
)
Once the tool wizard has opened (figure 5.1), choose the Pairwise Comparison table you would like to use.
Figure 5.1: Select a Pairwise Comparison table.
In the next dialog (figure 5.2), you can set the following parameters:
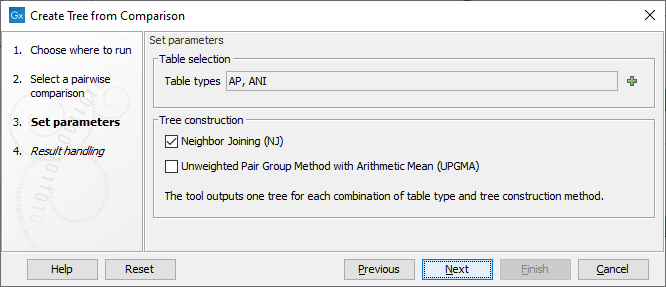
Figure 5.2: Select the table types and tree construction methods you would like to use for building trees.
- Table types The possible table types are extracted from the Pairwise Comparison table input. In the case of a Pairwise Comparison table obtained from Create Average Nucleotide Identity Comparison, these are: ANI (Average Nucleotide Identity) or AP (Alignment Percentage). If left empty, as it is by default, both types will be used.
- Tree construction method Neighbor Joining, well suited for trees with varying rates of evolution, and / or UPGMA, method that assumes a constant rate of evolution. Learn more about distance based reconstruction methods here: http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Distance_based_reconstruction_methods.html.
Note that the tool outputs a tree for each combination of table types and tree construction method. In addition, metadata from the Pairwise Comparison is automatically transferred to the tree. Sequence metadata containing taxonomy information is also added if this information was present in the inputs.
Learn more about visualizing trees here: http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Tree_Settings.html.
The Pairwise Comparison table input is either a distance or similarity matrix. The tool automatically detects the type of each table by checking the values on the diagonal: if the diagonal contains only zeros, then the table represents a distance matrix, otherwise it is a similarity matrix. From the table, a symmetric distance matrix d is calculated as follows:
d[i][j] = (t[i][j] + t[j][i]) / 2 if the table is a distance matrix,
d[i][j] = (1 - t[i][j] + 1 - t[j][i]) / 2 if the table is a similarity matrix,
where t[i][j] is the relative value (between 0 and 1) found in the table in row i and column j.
A tree is then created from the distance matrix d using the specified tree construction method. The tree is then generated such that the distance between two leaves (calculated as the sum of lengths of the branches connecting the leaves) is below 1, as the relative distance was used above. The branch lengths are then scaled so that the distance between two leaves reflects the absolute distance between them to match the entries in the table.
