Import high-throughput sequencing data
The Biomedical Genomics Workbench has dedicated tools for importing data from the following High-throughput sequencing systems.
- Illumina's Genome Analyzer, Nextseq, HiSeq and MiSeq
- PacBio
- Ion Torrent
- Complete Genomics (only processed data - master var and evidence files)
Importers for Roche 454 and SOLiD are also available in the Legacy Tools folder.
The reason for having dedicated tools for this is to standardize the data so that most downstream analyses and visualization of the data works seamlessly with all sequencing platforms. In case a sequence list was not imported with the right tool, it is possible to edit "Read Group" information in the "Element Info" view: choose from the drop-down menu the sequencing platform that was used to generate the data (figure 6.7) and click OK.
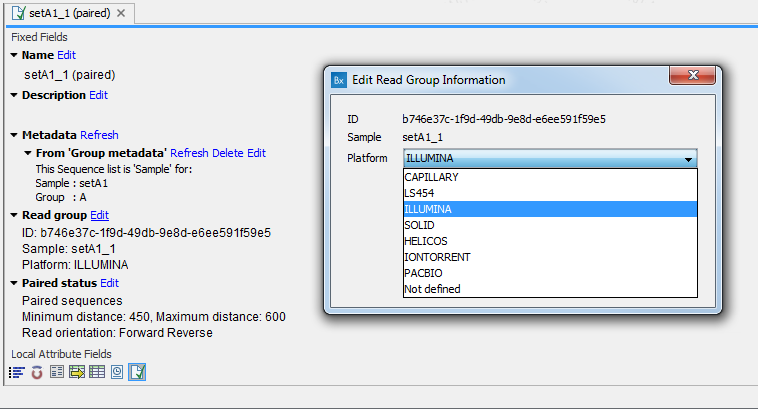
Figure 6.7: Editing the platform that was used to generate the data in the "Element Info" view.
In addition to these formats, mapped data in SAM/BAM format can also be imported.
Clicking on the Import (![]() ) button in the top toolbar will bring up a list of the supported data types as shown in figure 6.8.
) button in the top toolbar will bring up a list of the supported data types as shown in figure 6.8.
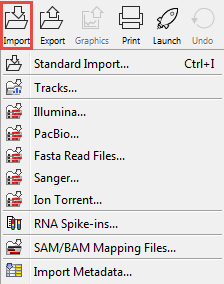
Figure 6.8: Choosing what kind of data you wish to import.
Select the appropriate format and then fill in the information as explained in the following sections.
Please note that alignments of Complete Genomics data can be imported using the Complete Genomics import.
Subsections
- Illumina
- PacBio
- Fasta read files
- Sanger sequencing data
- Ion Torrent
- Complete Genomics
- General notes on handling paired data
- SAM and BAM mapping files
