Analyze Expression Data and Upload Comparisons to IPA
The workflow Analyze Expression Data and Upload Comparisons to IPA takes expression data as input, and analyzes them using the tools in the RNA-Seq Analysis folder. It then submits the comparison to IPA using the Pathway Analysis tool.
The purpose of the workflow is to make it as easy as possible for the user to get from Sample to Insight. The user only has to provide expression data as input, and the workflow generates all available statistical analyses and data interpretation capabilities available via CLC Genomics Workbench and IPA.
Opened in the workflow editor, the workflows looks like this (see figure 4.6 below):
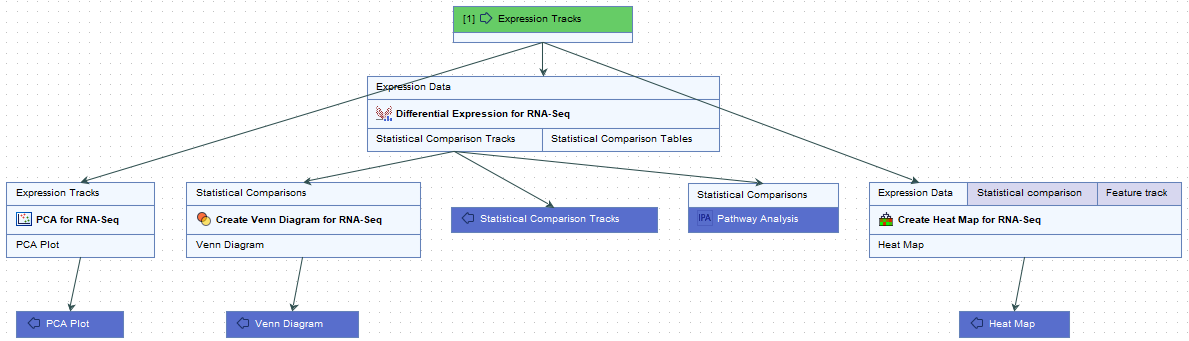
Figure 4.6: Layout of the Analyze Expression Data and Upload Comparisons to IPA workflow
The expression tracks are sent to three tools:
- Create Heat Map for RNA-Seq The tool creates a two dimensional heat map of expression values. Each column corresponds to one sample, and each row corresponds to a feature (a gene or a transcript). The samples and features are both hierarchically clustered.
- Differential Expression for RNA-Seq The tool performs a statistical differential expression test for a set of Expression Tracks. Its outputs are used as inputs for the IPA tool and for Create Venn Diagram for RNA-Seq (see below).
- PCA for RNA-Seq The tool creates a PCA plot, which is a projection of a high-dimensional dataset (where the number of dimensions equals the number of genes or transcripts) onto two of three dimensions. This helps in identifying outlying samples for quality control, and gives a feeling for the principal causes of variation in a dataset.
The outputs from the tools are saved in the chosen output folder for the workflow. The outputs from the Differential Expression for RNA-Seq tool are furthermore used for processing by these two tools:
- Pathway Analysis The tool uploads the comparisons to IPA. See Uploading data to IPA using the Pathway Analysis tool for details.
- Create Venn Diagram for RNA-Seq The tool makes it possible to compare two or more statistical comparison tracks. The Venn diagram comparison visualizes the overlap between the differentially expressed genes or transcripts in the selected statistical comparison tracks. The genes considered to be differentially expressed can be controlled by setting appropriate p-value and fold change thresholds.
Subsections
