Visualization of K-mer Tree for identification of common reference
The k-mer tree below (figure 13.17) includes 46 samples and 44 Salmonella genomes. To identify a candidate common reference genome, the tree was visualized using the radial tree topology setting. The common reference is usually chosen as the genome sharing the closest common ancestor with the clade of isolates under study in the k-mer tree. In this case, a reference (acc no NC_011083) located in the centre region of the tree was selected as a common reference candidate.
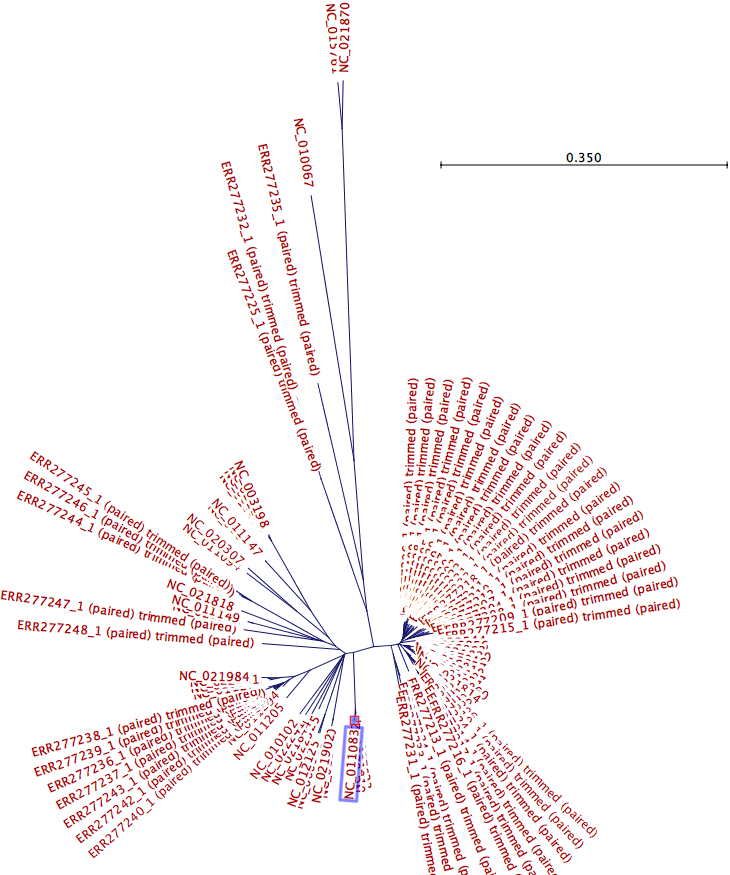
Figure 13.17: The created K-mer tree is visualized using the radial tree topology setting. The genome reference acc no NC_011083 situated in the center of the tree is selected as a common reference candidate.
If the sequence lists (samples and reference genomes) used as input for a k-mer tree contains metadata, the information will be used to decorate the tree.
The scale bar refers to the branch lengths within the tree.
Note that the information in the Taxonomy column of the sequence list needs to be following this format: "Kingdom; Phylum; Class; Order; Family; Genus; Species".
The metadata will also be made available in the K-mer tree table view, where you can manually edit entries in the metadata fields by right clicking on it in the tabular view of the Sequence List. If samples and reference genomes share metadata columns with the same header, these columns will be merged in both the K-mer tree table view and tree view.
Learn more about the overall Tree Settings, including how to decorate trees with metadata, here http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Tree_Settings.html.
