Set Up Gene Database
The Set Up Gene Database tool is meant for users who want to use their own list of resistance genes for resistance detection with the Find Resistance tool.
The tool can create new resistance gene lists/databases from a sequence list, or it can also be used to edit existing databases downloaded with the Download Database for Find Resistance tool. In the latter case, the tool imports resistance information to an existing resistance gene database and bundles the new metadata or overwrites the existing metadata sequence by sequence if this option is selected. This means that sequences that already have metadata and for which no new metadata is imported will keep the metadata; sequences with metadata for which new metadata is imported will be updated; sequences with no metadata will acquire new metadata.
To run the tool, go to:
Microbial Genomics Module (![]() ) | Databases (
) | Databases (![]() ) | Set Up Gene Database (
) | Set Up Gene Database (![]() )
)
In the first window (figure 15.8), select a new sequence list or an an existing database (with or without resistance genes information).
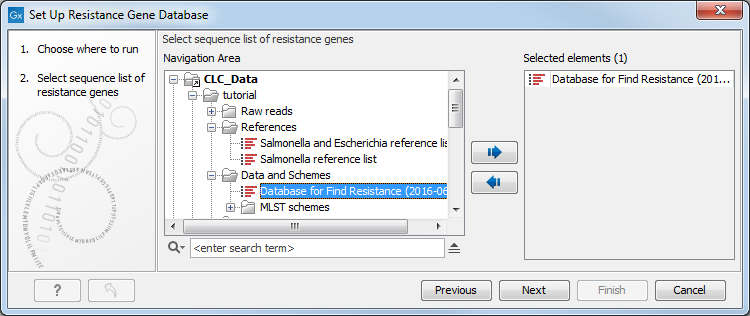
Figure 15.8: Select a sequence list or an existing resistance database you wish to modify.
In the second window (figure 15.9), select an Excel file (or a CSV file) containing the resistance information previously saved on your computer.
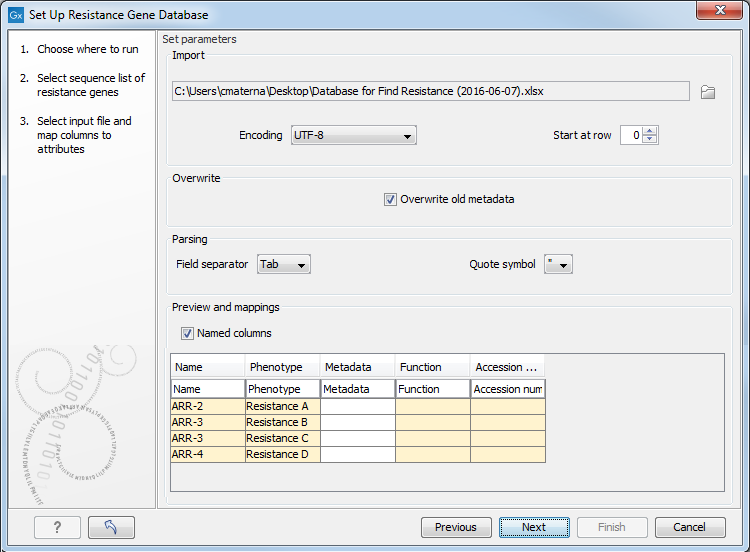
Figure 15.9: Select the file in which you saved the information you want to add to the sequence list.
There are 2 required columns in the input file: "Name" and "Phenotype" (i.e., resistance). You can add as many columns of metadata as necessary. The names given in the first row will be used as metadata categories.
Once imported, the information from the spreadsheet or CSV file will fill in the table included in the wizard window (figure 15.9), and the headers will take the names of the first row. It is still possible to edit the first row data at this point, thereby changing the names of the metadata categories. Leaving a first row field blank means that the metadata in that column will not be imported.
The required columns "Name" and "Phenotype", as well as the optional columns "Function" and "Accession number" from Resfinder are highlighted in yellow as they hold the information used subsequently by the Find Resistance tool.
Note that the option to overwrite previous metadata is selected by default but you can choose to deselect it in this window.
The tool will output a new database (or an updated database) containing the data included in the spreadsheet for all sequences whose name matched with the ones specified in the "Name" column of the spreadsheet. It is also possible to generate a report containing the number of genes in the database and the list of resistance phenotypes in the database (figure 15.10).
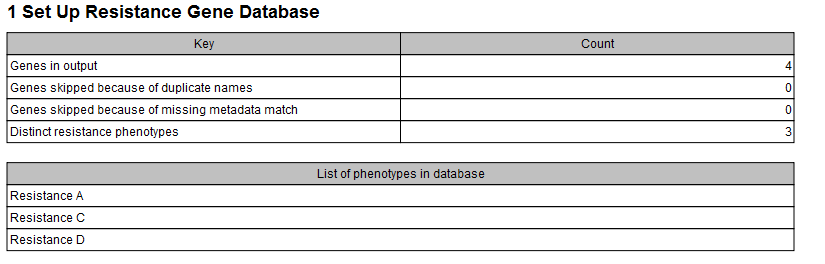
Figure 15.10: Example of a report produced by the Set Up Gene Database tool.
