Download Pathogen Reference Database
Download a collection of bacterial genomes representatives directly from RefSeq or NCBI Pathogen Detection Project (see http://www.ncbi.nlm.nih.gov/projects/pathogens/).
Microbial Genomics Module (![]() ) | Databases (
) | Databases (![]() ) | Download Pathogen Reference Database (
) | Download Pathogen Reference Database (![]() )
)
This will open the following wizard window (figure 15.4):
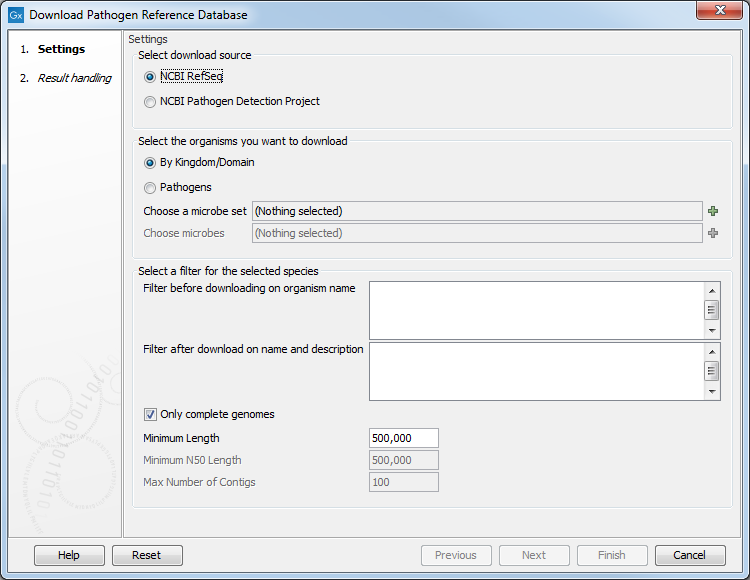
Figure 15.4: Downloading from the NCBI's RefSeq bacterial genomes database.
The settings are:
- Select download source RefSeq or Pathogen Project.
- Select the organism you want to download. This can be done By Kingdom/Domain, in which case you can choose a set from the predefined list (all archea, all bacteria, all fungi, all microbes, all plasmids, all viruses) or Pathogens, in which case you can choose an available set of microbes.
- Select a filter for the selected species: It is possible to filter before downloading on organism name, thereby potentially saving a lot of time for the download, or filter after download on name and description by defining one or more phrases to limit the output to those sequences which include at least one of these phrases in their description or their name.
- Only complete genomes
- Minimum Length: is by default set to 500,000 nt to favor bacterial genomes instead of plasmids.
- Minimum N50 length (the default value is set at 1,000,000 bp)
- Maximum number of contigs (the default value is set at 100) to ensure high enough quality of the database content
Specify a location to save the database. We recommend to create a folder where you can save all the databases and MLST schemes necessary to run some of the CLC Microbial Genomics Module tools.
The imported database includes a list of different bacterial genome sequences as well as the associated accession numbers (acc nos), descriptions, taxonomy and size of the sequences. In addition, each reference genome will be annotated with the following metadata (when available):
- serovar
- strain
- taxonomy
- sample collection date
- geographical location
- isolation source
- host
- host disease
- outbreak
- SRA run id
- SRA project id
Subsections
