Extract Regions from Tracks
The Extract Regions from Tracks tool facilitates specific extraction of mapped reads covering the particular regions specified by the annotation track, i.e., in this case the regions specified by an MLST scheme. The Extract Regions from Track tool is part of the template workflows Type a Known Species and Type among Multiple Species and the generated Track list enables visualization of the mapped reads per MLST loci. As the tool focuses on visualization of the mapping against the locus, the coverage up- and downstream of the loci does not reflect the actual coverage.
The tool is initiated by:
Microbial Genomics Module (![]() )| Typing and Epidemiology (beta) (
)| Typing and Epidemiology (beta) (![]() ) | Extract Regions from Track (
) | Extract Regions from Track (![]() )
)
The input file to the tool is a Reads Track, Genome Track or and/or Annotation Track (figure 10.50) while the specification of the regions to be extracted is specified by the NGS MLST Annotation Track (figure 10.51) generated when initially running the Identify MLST tool.
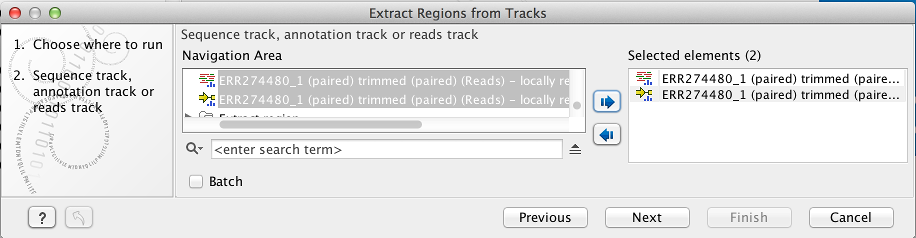
Figure 10.50: The first input file to the Extract Regions from Tracks tool is a Reads Track, Genome Track or and/or Annotation Track.
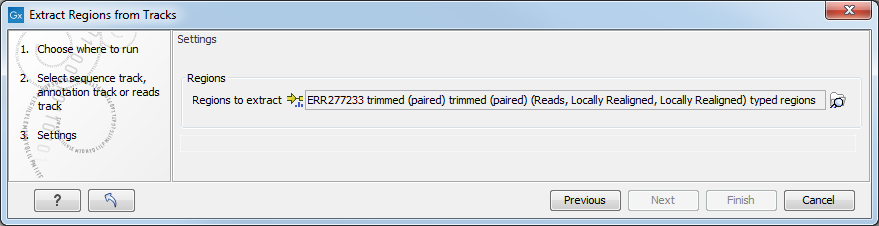
Figure 10.51: The second input file to the Extract Regions from Tracks tool is a NGS-MLST Annotation Track generated by the Identify MLST tool.
The extracted mapped reads is visualized in the track list shown in figure 10.52. Through the navigation tool at top right, it is possible to switch among the various MLST regions defined for the analyzed species.
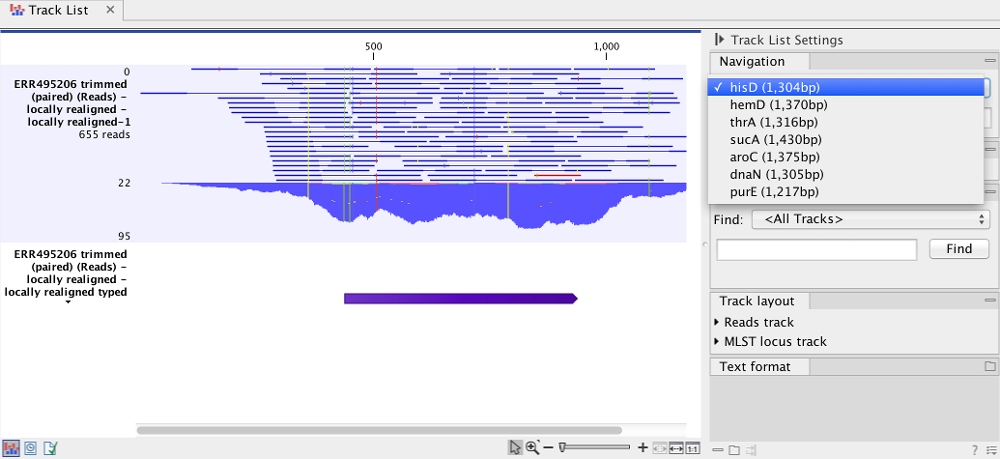
Figure 10.52: A track list showing the mapped reads covering a specific region defined by the species specific MLST scheme. At top right, the drop down list shows that the hisD region is currently visualized region.
