Parameters
The large gap mapper is started from the Toolbox:
Toolbox | Transcriptomics Analysis (![]() ) | Large Gap Mapper
) | Large Gap Mapper
After having specified the reads and the reference to which the reads should be mapped, the user must specify two parameters related to the mapped segments of a read (see figure 2.1:
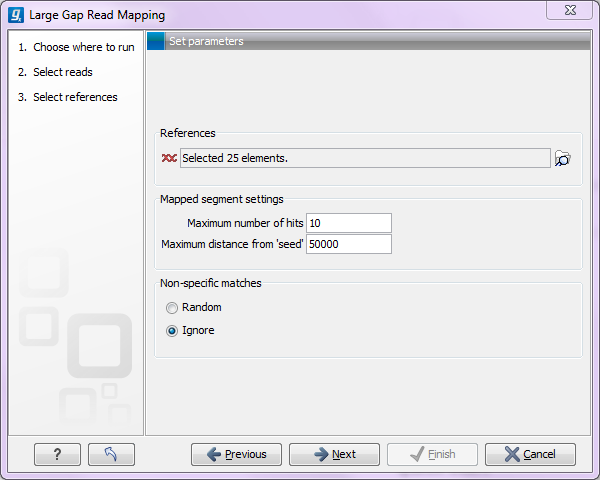
Figure 2.1: Specifying parameters for the large gap mapper.
- Maximum number of hits
- is the maximum number of hits that a segment is allowed to have in order for the read to be mapped. If, for a non-seed segment, this number is exceeded, the read is classified as unmapped. If it is not exceeded, all the multiple hit positions will be considered. If the seed makes up the full read it may map in up to 'Maximum number of hits' positions.
- Maximum distance from seed
- is the maximum distance allowed between seed and non-seed segments. Matches that are found further away from the seed that this value are discarded.
You can also specify whether non-specific matches should be distributed randomly or ignored.
Click Next to specify parameters related to the mapping quality. This is done in the Mapping settings step (see figure 2.2).
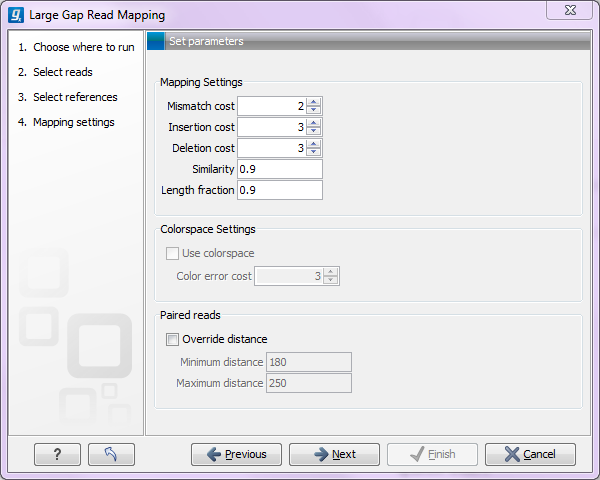
Figure 2.2: Specifying parameters for the large gap mapper.
Here, the can specify the mapping settings. We refer to the user manual of CLC Genomics Workbench for further detail (you can find the manual in the Help menu or at ). However, the the minimum similarity and length fractions need some more explanation: The similarity fraction is the required similarity between a mapped segment and the reference. This means that all segments must fulfill this requirement. Since segments can be as short as 17 bp, this threshold should not be set too strict (setting the threshold at 0.9 means that two errors for a segment of 17 bp would discard the match). The length fraction is the required fraction of the full read that should be mapped.
In addition to these user specified mapping settings, the large gap mapper requires that each mapped segment must comprise at least 10% of the read and must be of minimum length 17 bp (18 for color space).
Click Next to specify output options. In addition to the read mapping, the user can specify that a report on the mapping should be created, and that lists of unmapped and invalid mapped reads should be produced. The unmapped read list contains the reads which the large gap mapper was not able to map. The reads for which the large gap mapper was able to find a mapping, but for which the mappings of the segments where incompatible, are put in the invalid mapped reads list. The mappings of the segments of a read are incompatible if their positions are not consecutive along the reference, or if they do not have the same direction.
