Link Variants to 3D Protein Structure
This tool makes it possible to visualize variant consequences on 3D protein structures. It takes a variant track as input, and produces a new variant track as output, with two additional columns in the table view:
- Link to 3D protein structure: If a variant affects the amino acid composition of a protein, and a 3D structure of sufficient homology can be found in the Protein Data Bank (PDB), a link is provided in this column. Via the link, the structure can be downloaded and a 3D model and visualization of the variant consequences on the protein structure will be created.
- Effect on drug binding site: If any of the homologous structures found in PDB has a drug or ligand in contact with the amino acid variation, a link is provided in this column. Via the link, a list of drug hits can be inspected. The list has links for creating 3D models and visualizations of the variant-drug interaction.
In The variant table output it is described how to interpret the output in the variant table and how the tool finds appropriate protein structures to use for the visualizations, and in Create 3D visualization of variant and onwards it is described how the 3D models and visualizations are created.
|
Note: Before running the tool, a protein structure sequence database must be downloaded and installed using the Download 3D Protein Structure Database tool (see Download 3D Protein Structure Database). |
To run the tool, select:
Toolbox | Resequencing Analysis (![]() ) | Functional Consequences (
) | Functional Consequences (![]() ) | Link Variants to 3D Protein Structure (
) | Link Variants to 3D Protein Structure (![]() )
)
If you are connected to a server, you will first be asked where you want to run the analysis. In the next wizard step you will be asked for an input file. The Link Variants to 3D Protein Structure accepts variant tracks as input (see figure 25.78).
Figure 25.78: Select the variant track holding the variants that you would like to visualize on 3D protein structures.
Click on the button labeled Next. In the next wizard step, you must provide a CDS track and the reference sequence track (figure 25.79).
If you have not already downloaded a CDS and a reference sequence track, this can be done in the upper right corner of the Workbench by clicking on the button labeled Download and choosing Download Reference Genome Data (![]() ) (see Download reference genome).
) (see Download reference genome).
Figure 25.79: Select CDS and reference sequence.
Click on the button labeled Next, choose where you would like to save the data, and click on the button labeled Finish.
As output, the tool produces a new variant track, with two additional columns in the table view ('Link to 3D protein structure' and 'Effect on drug binding site' - figure 25.80). The default output view is the variant track. To shift to table view, click on the table icon found in the lower left corner of the View Area.
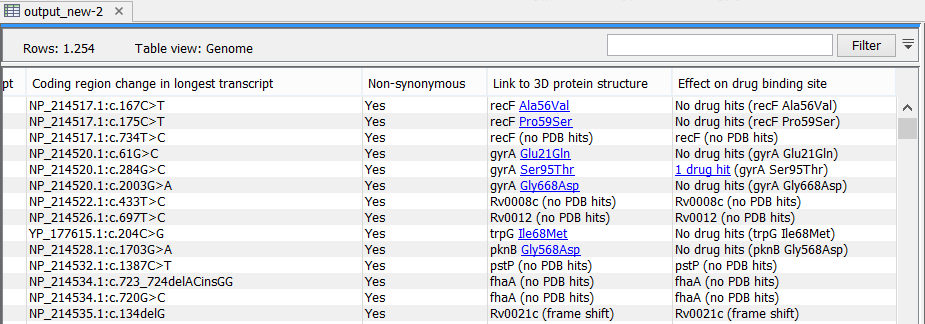
Figure 25.80: The variant table output.
Subsections
- The variant table output
- Create 3D visualization of variant
- Visualize drug interaction
- Protein coloring to visualize local structural uncertainties
- Ranking structures
- How a model structure is created
- How side chains are modeled
