Working with sequence lists
Sequence List elements contain one or more nucleotide sequences or one or more protein sequences. They are used as input to many tools, and are generated as output by many tools. Sequence lists can contain single end sequences or paired end sequences, but not a mixture of both. Handling paired data is described at the end of this section.
Much of the functionality available when working with sequence lists is also available when working with individual sequence elements. Shared functionality is described in working with sequences. This section describes options relevant only to sequence lists.
When open in the viewing area, there are several icons at the bottom providing access to different views of the data. The Side Panel view settings allow customization of what is shown and saving of customized settings.
In figure 15.7, the graphical view and Table view of a sequence list are open in linked views. With linked views, clicking in one view can lead to updates in the other view. For example, selecting a row in the Table view will cause the graphical view to update so the focus is on that sequence.
Right-click menus provide access to a wide range of features, with options differing depending on which view is open and where you click in it. Some actions are unique to a particular view, while others are available from more than one view. For example, sequence attributes can only be edited in the Table view but sequences can be added from the graphical view and Table view.
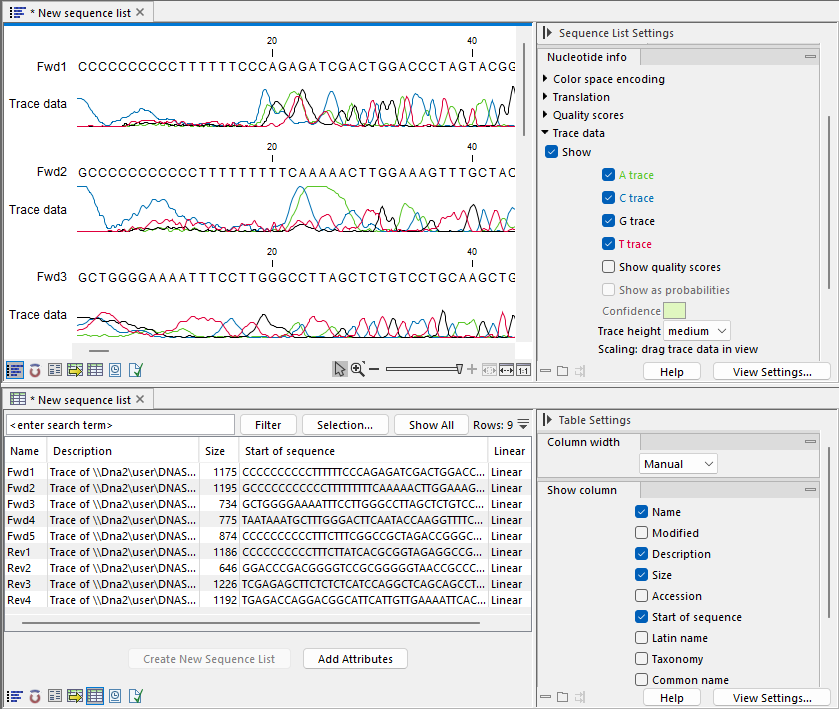
Figure 15.7: Two views of a sequence list are open in linked views, graphical at the top, and tabular at the bottom. Each view can be customized individually using settings in its Side Panel on the right.
Subsections
- Creating sequence lists
- Sequence List view
- Table view of sequence lists
- Annotation Table view of sequence lists
- Working with paired sequences in lists
