View Annotations in a table
The Annotation Table view (![]() ) lists the annotations in a table. Standard functionality for working with tables applies, see working with tables. Here, we highlight functionality of particular interest when working with annotations.
) lists the annotations in a table. Standard functionality for working with tables applies, see working with tables. Here, we highlight functionality of particular interest when working with annotations.
This view is useful for getting a quick overview of annotations, and for filtering so that only the annotations of interest are listed. From this view, you can edit and add annotations, export selected annotations to a GFF3 format file, and delete annotations. This functionality is described in more detail below.
To open the Annotation Table view:
- Select a sequence in the Navigation Area and right-click on the file name.
- Hover the mouse cursor over Show to see a list of options.
- Select Annotation Table (
 ).
).
Or, if the sequence is already open, click on the Show Annotation Table (![]() ) button at the bottom of the view. This will open a view similar to the one in figure 15.18.
) button at the bottom of the view. This will open a view similar to the one in figure 15.18.
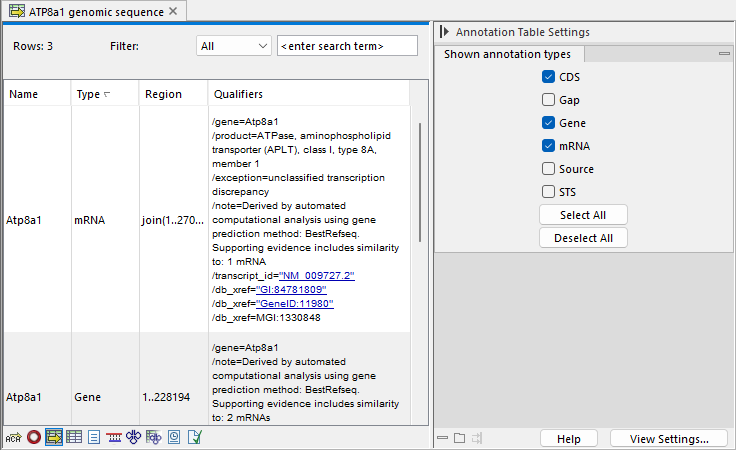
Figure 15.18: A table showing annotations on the sequence.
In the Side Panel you can show or hide individual annotation types in the table. E.g. if you only wish to see gene annotations, de-select all other annotation types.
Each row in the table is an annotation which is represented with the following information:
- Name
- Type
- Region
- Qualifiers
This information corresponds to the information in the dialog when you edit annotations and add annotations.
The Name, Type, and Region for each annotation can be edited simply by double-clicking, typing the change directly, and pressing Enter.
