Running analyses on the server
The tools available on the server are listed in the Introduction.
For more information on the tools, please see the manual for CLC Genomics Workbench at http://www.clcbio.com/usermanuals or click the Help (![]() ) button in the dialogs that are shown when you run the tools.
) button in the dialogs that are shown when you run the tools.
When you run the analyses you will be faced with an initial dialog with two choices:
- Workbench. This will run the analysis on your computer.
- Server. This will run the analysis on the server.
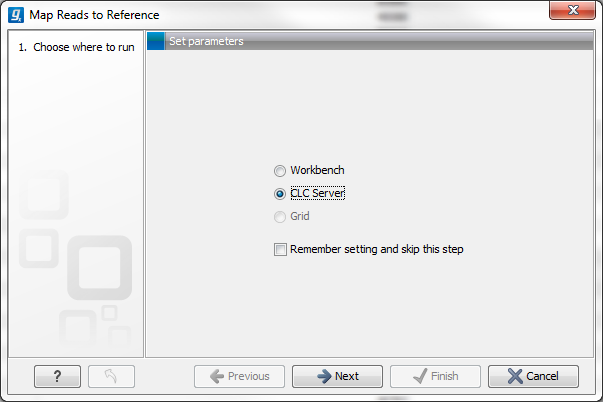
Figure 2.4: Selecting where to run the analysis.
Note that you can check the Remember setting and skip this step option to always use the selected option when the analysis dialog is shown. If you change your mind later on and want to switch, simply click Previous in the dialog and you will be able to change the setting.
Running the analyses on the server is identical to running the same analyses in the Workbench with three exceptions:
- You can only select data from the data locations connected to the server. This means that the Navigation Area is filtered to only show these locations.
- You have to save the result. In the last step, you can normally choose between Open and Save, but for the analyses performed on the server, you have to choose Save.
- When you click Finish, the analysis is performed on the server. This means that you can close the Workbench or disconnect the computer from the server, and the analysis will continue to run. When you log in after the analysis is done, you will get a notification in the Workbench.
Note that when using the import tool (Figure 2.5) for importing high-throughput sequencing data on the server (only for Genomics Workbench and Server), the first part of the import is uploading the data to the server and during this part, the Workbench has to maintain connection to the server. If you try to close the Workbench during upload, you will get a warning dialog. The upload runs as a local process in Processes tab in the lower left corner, and when the upload is done, a new process is started that has a server icon and runs on the server.
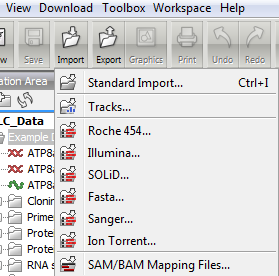
Figure 2.5: The import tool in the Workbench.
If you need help on the analyses themselves, please click the Help (![]() ) button in the dialog. That will take you to the relevant part of the user manual of the Workbench.
) button in the dialog. That will take you to the relevant part of the user manual of the Workbench.
