Download and configure reference data
The first time you open CLC Cancer Research Workbench you will be presented with the dialog box shown in figure 3.2, which informs you that data are available for download for either to the local or server CLC_References repository. If you check the "Never show this dialog again" then subsequently you will only be presented with the dialog box when updated versions of the reference data are available.
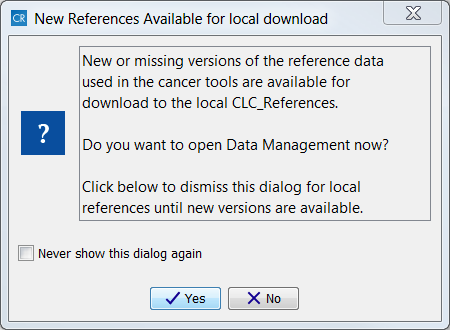
Figure 3.2: Notification that new versions of the reference data are available.
Click on the button labeled Yes. This will take you to the wizard shown in figure 3.3.
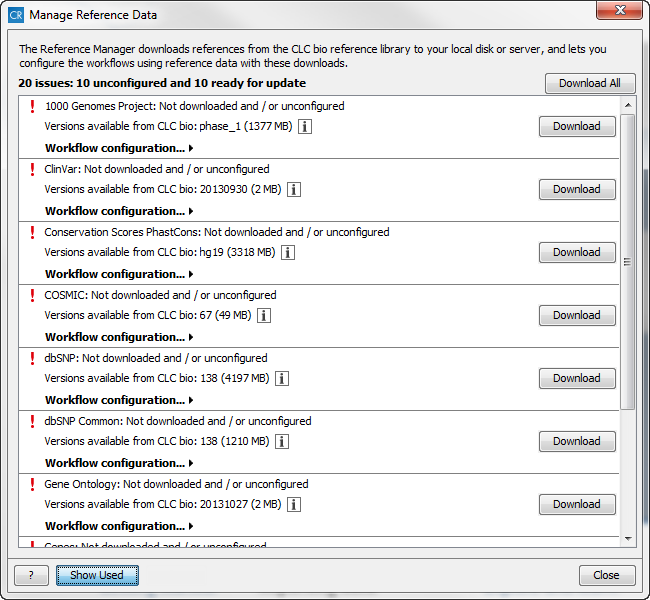
Figure 3.3: The Manage Reference Data wizard gives access to the reference data that are required to be able to run the ready-to-use workflows. The default view shows the references that are used in the workflows. With the "Show All" button the reference list can be expanded with additional (optional) reference data that you may find useful.
This wizard can also be accessed from the upper right corner of the CLC Cancer Research Workbench by clicking on Data Management (![]() ) figure 3.4.
) figure 3.4.
Figure 3.4: Click on the button labeled "Data management" to open the "Manage Reference Data" dialog where you can download and configure the reference data that are necessary to be able to run the ready-to-use-workflows.
The "Manage Reference Data" wizard gives access to all the reference data that are used in the ready-to-use workflows. From the wizard you can download and configure the reference data. A button labeled "Show All" at the bottom of the dialog can be used to expand the list with additional reference data that are not required for any of the workflows (e.g. Gene Ontology). Rather these extra reference data have been provided as an extra service for those of our users who would like to include information from these databases in the data analyses.
Icons are used in the "Manage Reference Data" wizard to give a quick overview of
the current status of each reference: "Not downloaded and / or unconfigured",
"Workflows use different versions" or "Selected version is inconsistent / not
fully downloaded" references are marked with a red exclamation mark
(![]() ), references that are "Up to date and configured" are
marked with a green check mark (
), references that are "Up to date and configured" are
marked with a green check mark (![]() ), and when a new
version of a reference data set is available, you will see a green mark labeled "New"
(
), and when a new
version of a reference data set is available, you will see a green mark labeled "New"
(![]() ).
).
Guide to the "Manage Reference Data" wizard:
- In the upper part of the wizard you can find:
- A small descriptive text
- An indication of how many issues you have, how many of these are "unconfigured issues", and how many are reference data that are "ready for update".
- The button labeled Download All, which can be used to download all reference data that are shown in the wizard. This is the case the first time you use the "Download All" button. Subsequently, only reference data where a newer version is available, will be downloaded. If you have selected "Show All" (the "Show All" button is found at the bottom of the wizard), all reference data will be downloaded (including "Gene Ontology"). If you have selected "Show Used", only the reference data that are used in the ready-to-use workflows will be downloaded.
- The central area of the wizard:
- Lists all available references data. After the reference name, a small note shows the status of the reference (see figure 3.5), which can be:
- Not downloaded and / or unconfigured (
 )
)
- Workflows use different versions (
 )
)
- Selected version is inconsistent / not fully downloaded (
 )
)
- Up to date and configured (
 )
)
- New version available (
 )
)
- Not downloaded and / or unconfigured (
- When a new version is available (
 ), it is stated
in a parenthesis whether it is for your local disc, for the server, or local and
server (see figure 3.5).
), it is stated
in a parenthesis whether it is for your local disc, for the server, or local and
server (see figure 3.5).
- If a version is inconsistent / not fully downloaded
(
 ), it will be stated in parenthesis whether it is
the local or server version (or both). Check the process tab for running or
suspended download processes. Please wait for all of these to finish. If the
data is inconsistent, even after all downloads have finished, it is likely that
you ran out of disk space, or the download or import was somehow stopped
prematurely.
), it will be stated in parenthesis whether it is
the local or server version (or both). Check the process tab for running or
suspended download processes. Please wait for all of these to finish. If the
data is inconsistent, even after all downloads have finished, it is likely that
you ran out of disk space, or the download or import was somehow stopped
prematurely.
In this case, you can "Delete" the reference, and try downloading it again. - In the unlikely event that a reference has the mark Workflows use different
versions (
 ), the Workbench has discovered that two
or more installed workflows use different versions of a reference, and is unable
to determine which should be used. Please select the correct version from the
drop down menu and click "Use Reference" to solve this. See Workflow
configuration below for more information on configuring workflows.
), the Workbench has discovered that two
or more installed workflows use different versions of a reference, and is unable
to determine which should be used. Please select the correct version from the
drop down menu and click "Use Reference" to solve this. See Workflow
configuration below for more information on configuring workflows.
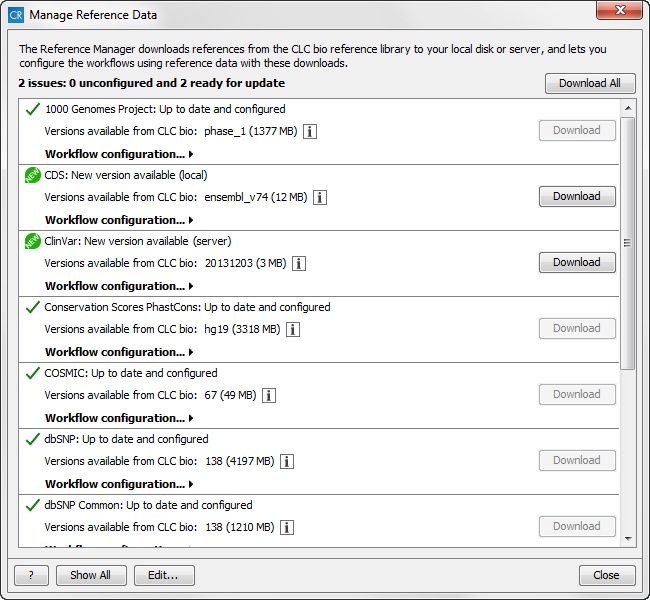
Figure 3.5: The Manage Reference Data wizard lists the reference data. Three different icons are used to mark the status of the reference. - Under the reference used, you can find info about the reference version
(Versions available from CLC bio) and the size of the reference data. By
clicking on (
 ) you can see the legal notice and
license information for this particular reference data set (see
figure 3.6).
) you can see the legal notice and
license information for this particular reference data set (see
figure 3.6).
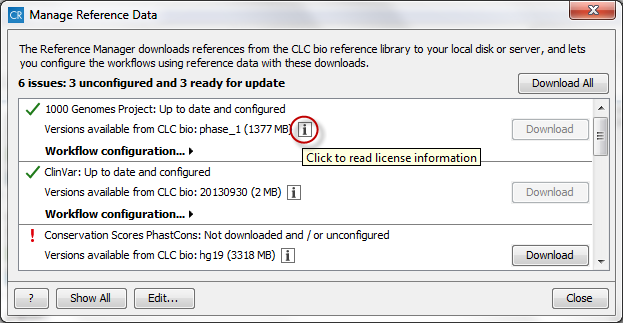
Figure 3.6: Click on the info button to see the legal notice and license information. - The button labeled Download can be used to download the reference data
individually. When you click on the button labeled Download, a wizard
appears with a message informing you that the selected reference data are now
being downloaded (figure 3.7).
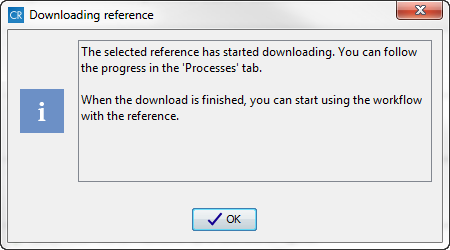
Figure 3.7: The Downloading Reference wizard informs you about that data is being downloaded.After the reference data have been downloaded the icon changes to a green check mark for those of the databases that only contain one reference data file.
- The boldface text Workflow configuration can be expanded to reveal additional options. When unfolded, you can see which version of the reference is being used, and which of the ready-to-use workflows use this reference. In addition, three buttons appear:
- Use Reference When the reference data have been downloaded,
the workflows will automatically be configured with all the reference
data available. The drop down "Select Version" allows you to change
between the downloaded versions, and pressing "Use Reference" will
update the installed workflows to use this specific version for the
selected reference.
However, for references like the "1000 Genomes Project" and "HapMap" databases,
which contain more than one reference data file3.3, you have to
specify which reference data to use. This is what the "Use Reference"
option allows you to do. Select the reference data by clicking on the
data you want to use. If you want to select more than one population,
hold down the Ctrl key while selecting data files.
When you have selected the population that you want to use for your data analyses, click on the button labeled OK. Your workflow will now be configured with the reference data for the population(s) that you have selected. Please note that you have to do this for both the "1000 Genomes Project" and for the "HapMap" reference data. See figure 3.8.
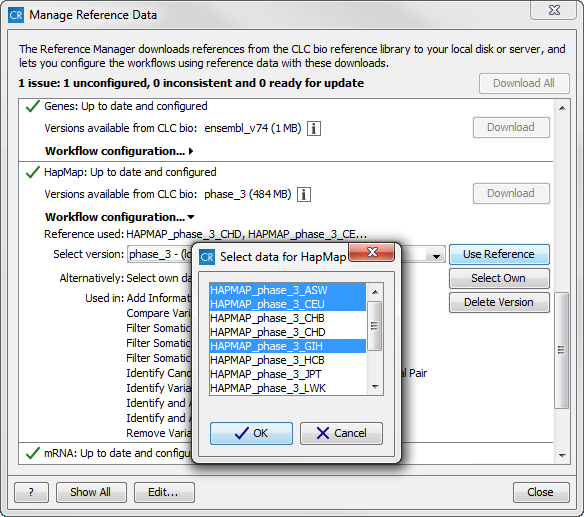
Figure 3.8: Select the population variant track that you want to use in your ready-to-use workflows. - Delete Version With this button all users are capable of deleting locally installed reference data, whereas only administrators are capable of deleting reference data installed on the server. This can be used if you suspect that a downloaded reference is corrupt, and needs to be re-downloaded, or if you need to clean up space, e.g. locally.
- Use Own File allows you to use your own reference data. The
data type and number of files to select will be restricted to match the
reference. This is useful when you have your own version of the
reference data that you would like to use rather than the data made
available to download directly into the Workbench. If you want to switch
back to using the downloaded references, you must use the "Use
Reference" again.
- Use Reference When the reference data have been downloaded,
the workflows will automatically be configured with all the reference
data available. The drop down "Select Version" allows you to change
between the downloaded versions, and pressing "Use Reference" will
update the installed workflows to use this specific version for the
selected reference.
However, for references like the "1000 Genomes Project" and "HapMap" databases,
which contain more than one reference data file3.3, you have to
specify which reference data to use. This is what the "Use Reference"
option allows you to do. Select the reference data by clicking on the
data you want to use. If you want to select more than one population,
hold down the Ctrl key while selecting data files.
- Lists all available references data. After the reference name, a small note shows the status of the reference (see figure 3.5), which can be:
- At the bottom of the wizard you can find:
- A button with a question mark. This is the "help" button that links to the section in the CLC Cancer Research Workbench reference manual that describes the "Manage Reference Data" button.
- A button labeled "Show All" (or "Show Used"). With this button you can choose whether you only want to see the reference data that is being used in the ready-to-use workflows, or if you want to see all available reference data. Please note that if you choose to use the "Download All" function, you will download the references that are shown in the wizard. This means that if you have selected "Show Used" you will only download the reference data that is being used in the workflows.
- A button labeled "Close". Click on this to close the wizard.
If you are connected to a CLC Server you will be asked where you want to save the downloaded reference data, to your Workbench or your Server when you click on the button labeled Download or Download All. See figure 3.9. You will see this dialog the first time you download data. After this the dialog will appear only in situations where both the Local and Server version need updating. If a new version is found with respect to only Local or Server, the data will automatically be downloaded to that location.
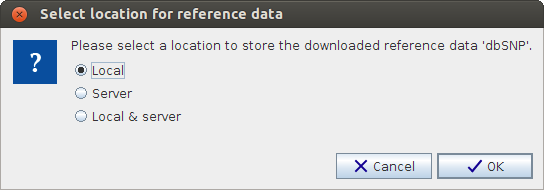
Figure 3.9: Select where to save the downloaded reference data. Please be aware that the total size of all reference data (in April 2014) is about 12 GB when compressed. It can take some time to download all reference data. When unzipped the size of all the reference data, when the compressed size was about 12 GB is about 75 GB.
When the reference data have been downloaded, the workflows will automatically be configured with the reference data. However, in some cases reference data are available from different population subgroups. This is the case for HapMap and the 1000 Genomes Project. Three letter codes are used to specify the population that the different reference data origin from (e.g. ASW = American's of African Ancestry in SW USA). For the phase 3 HapMap population codes, please see http://www.sanger.ac.uk/resources/downloads/human/hapmap3.html and for the 1000 Genomes Project see http://www.ensembl.org/Help/Faq?id=328.
Whenever workflows use reference data that are available from more than one population, the workflow will initially be configured with all the populations being available, and which population to use in the workflow will then need to be specified by the user in one of the wizard steps that appear when starting the workflow. How to configure your workflow with the right population is described in Download and configure reference data.
Figure 3.10 shows the CLC_References folder. If you open the folders holding the reference data, you can see that different populations are available for HapMap and the 1000 Genomes Project.
Figure 3.10: For the 1000 Genomes Project and HapMap reference data, data are available from different populations. For these two databases the user must manually specify the relevant population to be used in the workflows. If the user choose not to select a population manually, the workflow will use a randomly selected population.
Footnotes
- ... file3.3
- In some cases, reference data are available from different population subgroups. This is the case for HapMap and the 1000 Genomes Project. Three letter codes are used to specify the population that the different reference data origin from (e.g. ASW = American's of African Ancestry in SW USA). For the phase 3 HapMap population codes, please see http://www.sanger.ac.uk/resources/downloads/human/hapmap3.html and for the 1000 Genomes Project see http://www.ensembl.org/Help/Faq?id=328. Figure 3.10 shows the CLC_References folder. You can see that different populations are available for HapMap and the 1000 Genomes Project.
