Output from the Identify and Annotate Variants (WES) workflow
The Identify and Annotate Variants (WES) tool produces several outputs.
Please do not delete any of the produced files alone as some of them are linked to other outputs. Please always delete all of them at the same time.
A good place to start is to take a look at the mapping report to see whether the coverage is sufficient in the regions of interest (e.g. > 30 ). Furthermore, please check that at least 90% of the reads are mapped to the human reference sequence. In case of a targeted experiment, please also check that the majority of the reads are mapping to the targeted region.
Next, open the Genome Browser View file (see figure 6.46).
The Genome Browser View includes a track of the identified annotated variants in context to the human reference sequence, genes, transcripts, coding regions, targeted regions, mapped sequencing reads, clinically relevant variants in the ClinVar database as well as common variants in common dbSNP, HapMap, and 1000 Genomes databases.
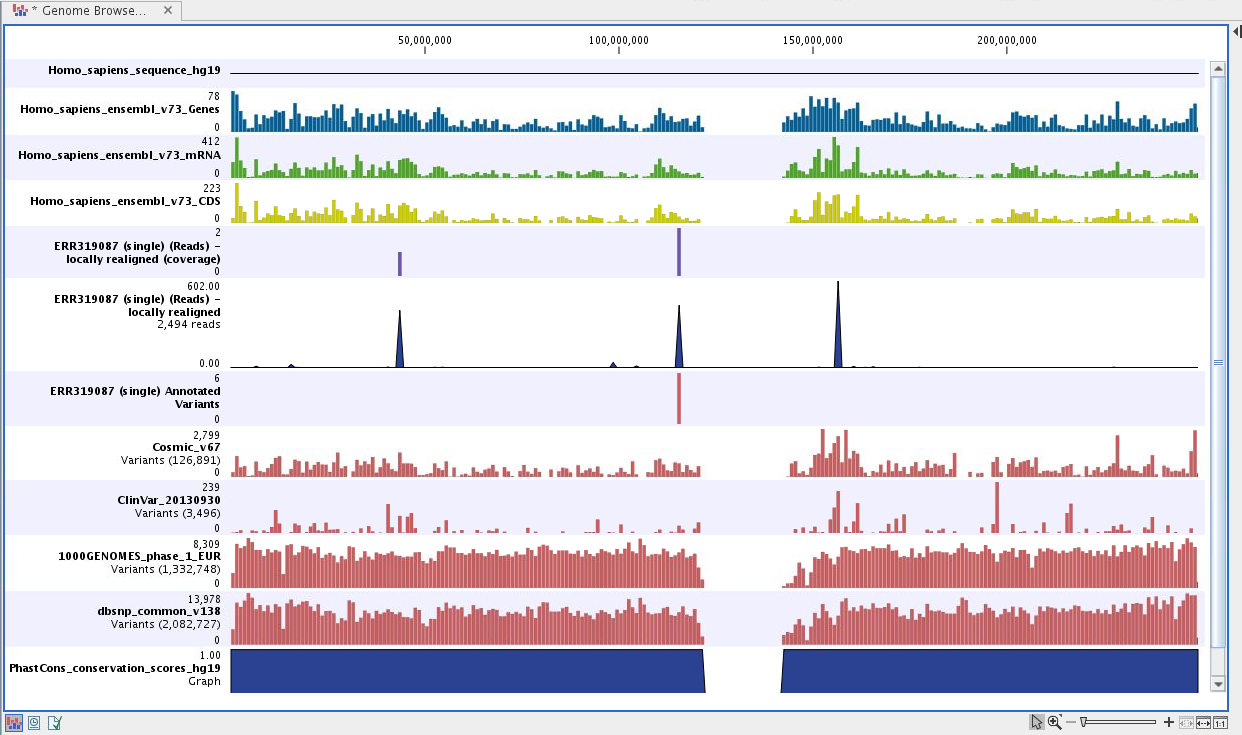
Figure 6.46: Genome Browser View to inspect identified variants in
the context of the human genome and external databases.
To see the level of nucleotide conservation (from a multiple alignment with many vertebrates) in the region around each variant, a track with conservation scores is added as well.
By double-clicking on the annotated variant track in the Genome Browser View, a table will be shown that includes all variants and the added information/annotations (see figure 6.47).
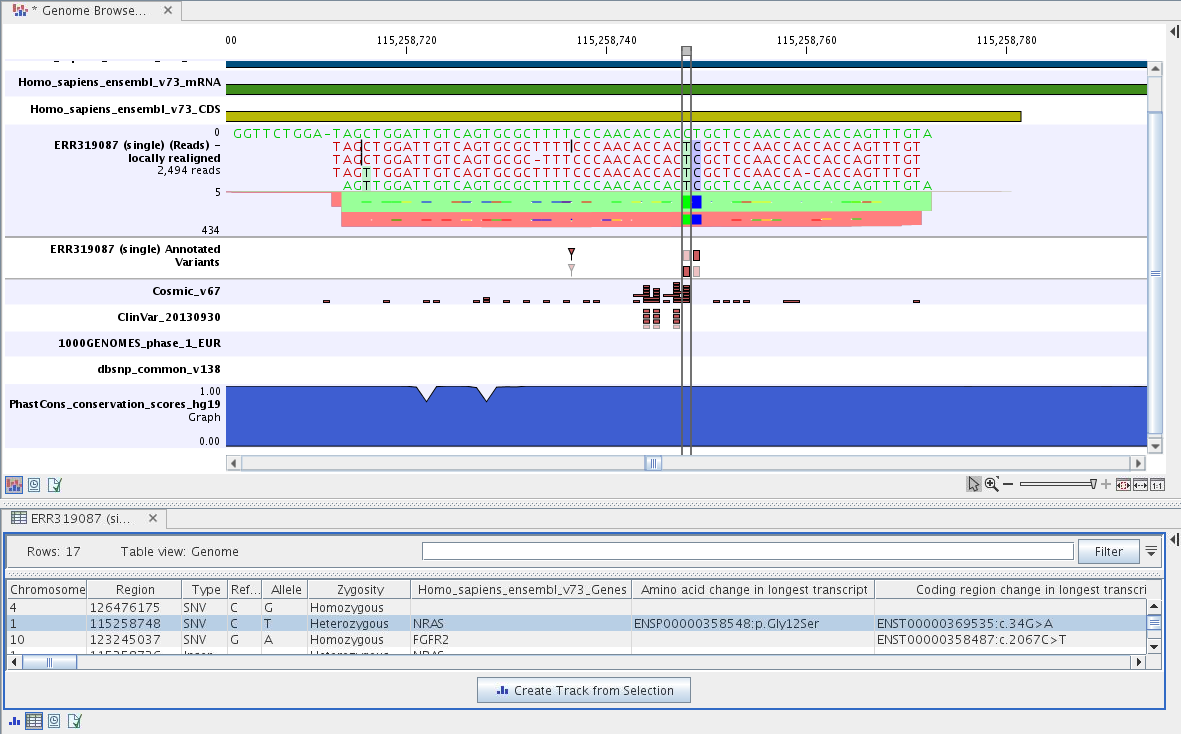
Figure 6.47: Genome Browser View with an open track table to
inspect identified somatic variants more closely in
the context of the human genome and external databases.
The added information will help you to identify candidate variants for further research. For example can common genetic variants (present in the HapMap database) or variants known to play a role in drug response or other clinical relevant phenotypes (present in the ClinVar database) easily be seen.
Not identified variants in ClinVar, can for example be prioritized based on amino acid changes (do they cause any changes on the amino acid level?). A high conservation level on the position of the variant between many vertebrates or mammals can also be a hint that this region could have an important functional role and variants with a conservation score of more than 0.9 (PhastCons score) should be prioritized higher. A further filtering of the variants based on their annotations can be facilitated using the table filter on top of the table.
If you wish to always apply the same filter criteria, the Create new Filter Criteria tool should be used to specify this filter and the Identify and Annotate Variants (WES) workflow should be extended by the Identify Candidate Tool (configured with the Filter Criterion). See the reference manual for more information on how preinstalled workflows can be edited.
Please note that in case none of the variants are present in ClinVar or dbSNP, the corresponding annotation column headers are missing from the result.
In case you like to change the databases as well as the used database version, please use the "Data Management".
