Create a custom Reference Data Set
The Reference Data Sets also contain a Create Custom Set ... button that allows you to create your own set of reference data starting from an existing data set
Clicking on this button will open a window (figure 4.8) where you can change the name of the new data set, the organism it represents, the chromosomal extension, and the annotation types used.
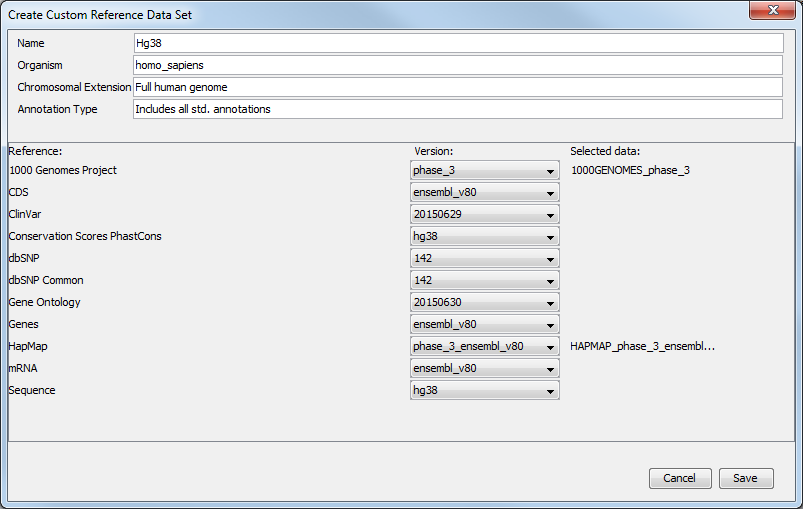
Figure 4.8: Select the reference data elements you want to add to you custom reference data set.
For each type of reference, a drop-down menu allows you to choose from the different versions available, as well as a custom option that allows you to import database and sequences saved in your Navigation Area (figure 4.9). This is useful when you have your own version of the reference data that you have imported in the workbench and that you would like to use rather than the currently available Reference Data Sets. The customs data sets are saved under the Custom Reference Data Sets tile. Do not forget to click on the button Apply if you wish to use this set for your workflows.
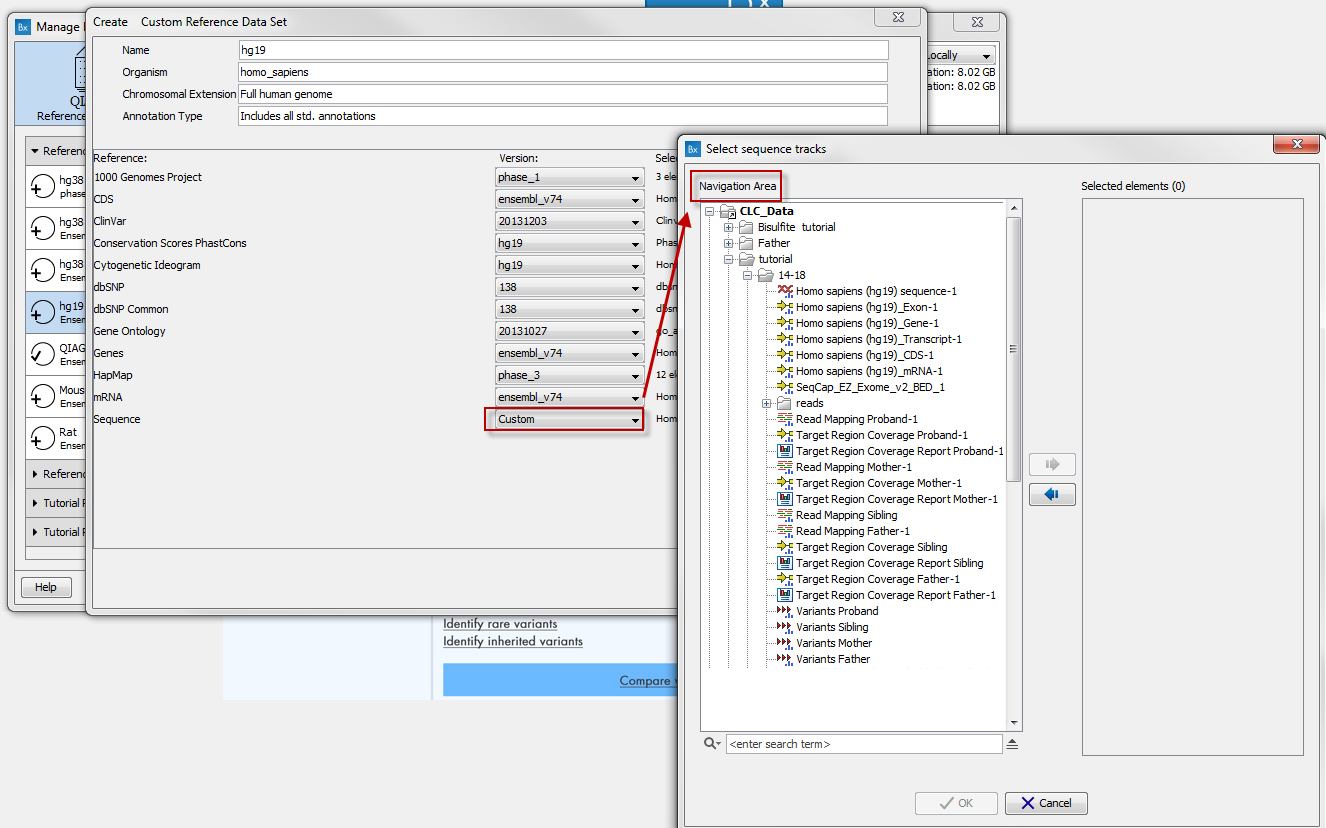
Figure 4.9: Select data from your Navigation Area to create your own custom Reference Data Set.
For references like the "1000 Genomes Project" and "HapMap" databases which contain more than one reference data file, the workflow will initially be configured with all the populations being available and you will be able to specify which reference data to use in the workflow wizard directly. But you can also modify a pre-existing Reference Data Set to contain only the population you want to work with. In the Data Management wizard, select the Reference Data Set you are interested in, click on Create Custom Set. Select the version of the 1000 genomes or Hapmap database you wish to work with (figure 4.10).
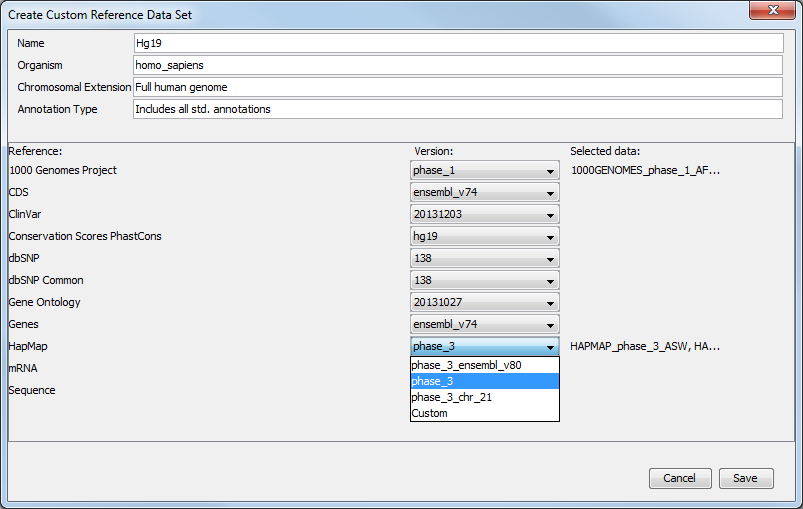
Figure 4.10: Select the version of the 1000 genomes or Hapmap database you want to work with, or select the option "custom".
A pop-up window will open where you can select the population you want to work with. Alternatively, click on the option "custom" in lieu of version and choose from the CLC_References folder the population of your choice (figure 4.11).
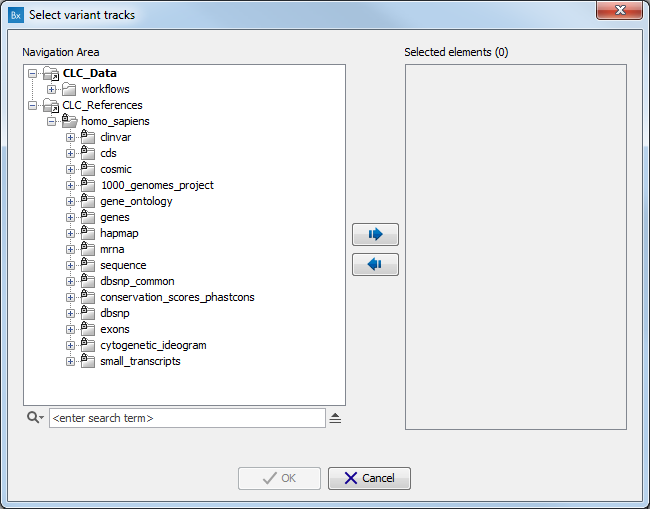
Figure 4.11: Choosing the option "custom" allows you to choose your reference from the Navigation Area..
Three letter codes are used to specify the population that the different reference data origin from (for example ASW = American's of African Ancestry in SW USA). For the phase 3 HapMap population codes, please see http://www.sanger.ac.uk/resources/downloads/human/hapmap3.html and for the 1000 Genomes Project see http://www.1000genomes.org/category/frequently-asked-questions/population.
Note: Custom reference data sets specific to the workbench on which they are created, and will not appear in other workbenches connected to the same server.
