Detect Fusion Genes
The Detect Fusion Genes tool can be found in the Toolbox here:
Tools | QIAseq Panel Expert Tools | QIAseq RNAscan Panel Expert Tools | Detect Fusion Genes
The Detect Fusion Genes tool takes an RNA-seq read mapping as input (figure 4.8).
Figure 4.8: Select an RNA-seq read mapping.
In the next dialog (figure 4.9), specify the reference sequence, gene and mRNA track that are saved in the CLC_References folder of the Navigation Area when downloading the QIAseq RNAscan Panels hg38 Reference Data Set. It is possible - but optional - to add a CDS or primer track to run the analysis.
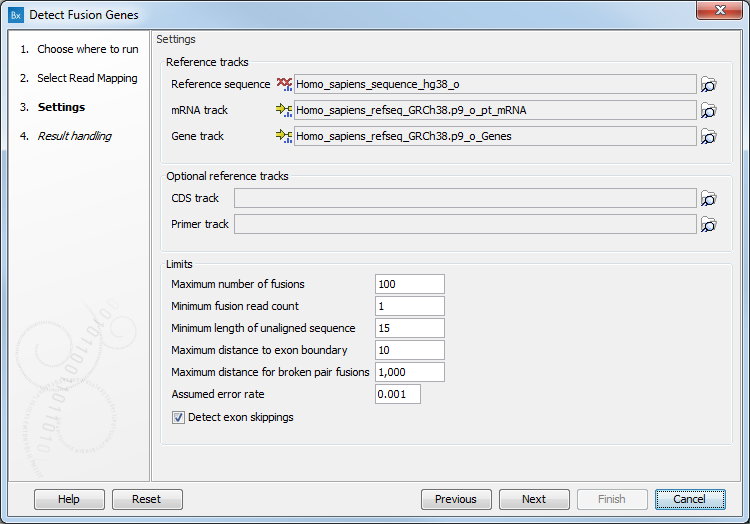
Figure 4.9: Specify references and parameters for the detection.
The additional parameters to set are:
- Maximum number of fusions: The maximum number of putative fusions that will be evaluated.
- Minimum fusion read count: This value is used to calculate Z-score and p-value, by subtracting that number from the total read count before doing the statistics.
- Minimum length of unaligned sequence: Only unaligned ends longer than this will be used for detecting fusions.
- Maximum distance to exon boundary: Reads must break within this distance of an exon boundary to consider the read and exon compatible.
- Maximum distance for broken pairs fusions: The algorithm uses broken pairs to find additional support for fusion events. If a pair of reads originally mapped as a broken pair, but would not be considered broken if mapped across the fusion breakpoints (because the two reads in the pair then get close enough to each other), then that pair of reads supports the fusion event as "fusion spanning reads". The "Maximum distance for broken pairs fusions" parameter specifies how close to each other two broken pairs must map across the fusion breakpoints in order for them to be considered fusion spanning reads. This is usually set to the maximum paired end distance used for the Illumina import of reads.
- Assumed error rate: Value used to calculate Z-score and p-value.
- Promiscuity threshold: Minimum number of different fusion partners to consider a gene promiscuous. Fusions with a promiscuous gene will get the annotation "Promiscuous" in the Filter column.
- Detect exon skippings: Check this option to consider same-gene fusions, i.e., exon-skipping fusions. Note however that same-gene fusions where the 5' breakpoint is downstream of the 3' breakpoint will not be considered.
In the Result handling dialog, it is possible to choose to output a report with unaligned ends information (figure 4.10):
- Unaligned ends: number of found unaligned ends.
- Mapped unaligned ends: number of unaligned ends which could be mapped
- Unmapped unaligned ends: number of unaligned which could not be mapped.
- Discarded base breakpoints: when two transcripts of the same gene overlap so that two breakpoints are found next to each other, one of them will be discarded.

Figure 4.10: Unaligned ends report.
The tool otherwise generates a read mapping and a fusion track (see the details of the fusion track in Output from the Detect QIAseq RNAscan Fusions workflow section.
