MAFFT
MAFFT is a multiple sequence alignment tool [Katoh et al., 2002]. MAFFT uses a number of heuristics to speed up the alignment process, making it suitable for aligning large numbers of sequences. See https://mafft.cbrc.jp/alignment/software/ for more information.
MAFFT alignment options
- Gap cost. Increasing the gap cost will favor fewer gaps in the alignment.
- Gap extension cost. Increasing the gap extension cost will favor shorter gaps in the alignment.
- Adjust direction. When enabled, nucleotide sequences are reverse-complemented as needed. In the output alignment, reverse-complemented sequences will have "-RC" appended to their name. Enabling this option may increase run time.
- Reference sequence or alignment. Optionally select a reference sequence or an alignment to which the input will be aligned.
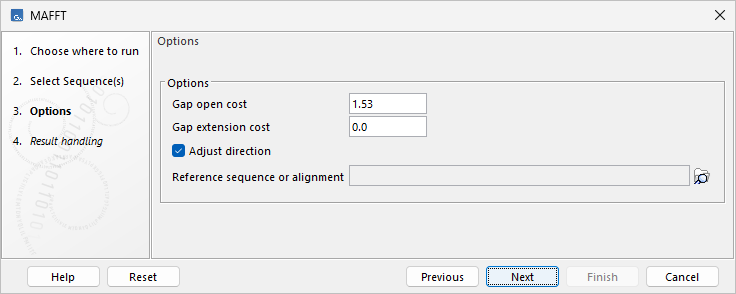
Figure 2.4: MAFFT alignment options.
Note: When running MAFFT in CLC Workbench, the tool automatically selects the appropriate alignment strategy based on the input size. For amino acid sequences, it uses the BLOSUM62 scoring matrix, and for nucleotide sequences, it applies the 200PAM scoring matrix. For general information on scoring matrices see Bioinformatics explained: Scoring matrices.
MAFFT output options
MAFFT produces a single alignment as output and, optionally, the constructed guide tree.
- Output guide tree. Output the guide tree together with the alignment. This option is not available when a reference sequence or alignment has been provided.
If "Adjust direction" was enabled, any reverse-complemented sequences have "-RC" appended to their name in the alignment and in the guide tree.
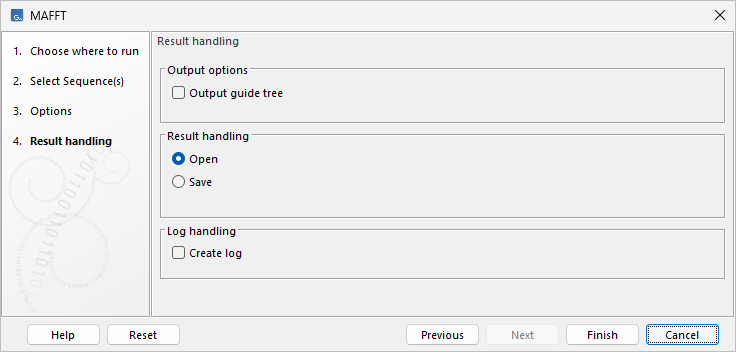
Figure 2.5: MAFFT output options.
