Annotate and filter from variant database
Comparison with variant databases is a key concept when working with
resequencing data. The CLC Genomics Workbench provides two tools for facilitating this
task: one for annotating your experimental variants with information from
database variants (e.g. adding information about phenotypes like cancer
associated with a certain variant allele), and one for filtering your
experimental variants based on this information (e.g. for removing common
variants).
In order to do this, you will have to import or download a file that is recognized as a variant file (see Import tracks from file and Download reference genome).
This section will use the filter tool as an example, since the core of the tools are the same:
Toolbox | Resequencing (![]() ) | Annotate and Filter | Filter against Variant Database
) | Annotate and Filter | Filter against Variant Database
This opens a dialog where you can select a variant track (![]() ) with experimental data that should be filtered.
) with experimental data that should be filtered.
Clicking Next will display the dialog shown in figure 26.18
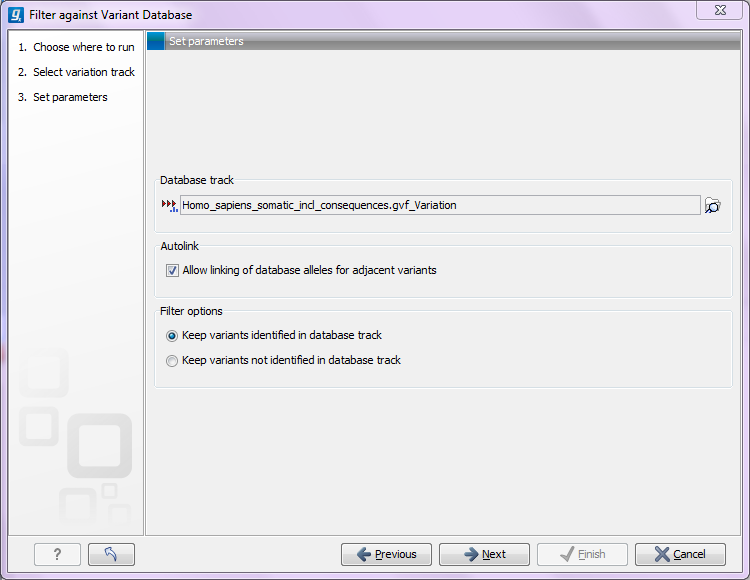
Figure 26.18: Specifying a variant database to filter against.
Select (![]() ) a database to compare against. The tool will then compare
the variants provided in the input track with the ones reported in the database
track and evaluate whether it is known in the database.
) a database to compare against. The tool will then compare
the variants provided in the input track with the ones reported in the database
track and evaluate whether it is known in the database.
- If the position is the same and all alleles of the variant are identified the database track, it is a known variant.
- If the position is the same and only one out of three alleles of the variant are identified in the database track, it is an unknown variant.
- If the position is the same, but the alleles are not the same as in the database, it is an unknown variant.
- If the position is not known in the database, it is an unknown variant.
The option to Allow linking of database alleles for adjacent variants comes into play for MNVs or larger InDels where the CLC Genomics Workbench reports one variant and the database may have this split up into several SNVs or InDels (some databases do not include linked information about SNVs or InDels). As an example, you may have a variant like this AC>GT. The database track may have the same variant represented as two entries: A>G and C>T. In this case, it can be difficult to compare the data, since the database track does not hold information about how the individual SNVs are linked (i.e. come from the same alleles). So even though the variant can be found by splitting it up, the database comparison tool has to link the database variants together to compare them. Since this cannot be based on data, it is an option that you can choose to deselect. This would mean missing potential matches with the database, but on the other hand there will no assumptions about linkage.
The annotation version of the tool will create a new track with all the experimental variants including added information about known variants. The filtering version as shown in figure 26.18 allows you to create a new track by Keeping variants found in database track or by Keeping variants not found in database track. When keeping variants found in the database track, these will also be annotated.
In case only part of an identified MNV or InDel is present in the database (e.g. In case of AT->GC only A->G is present in the database, but T->C is not) the tool will add an annotation with name Partial Match and the name of the database, which includes part of the variant. Please note that only submatches to database variants are annotated as partial matches and not the other way around. So in case A->G has been identified as variant in the sample, but only AT->GC is present in the database, this will not reported as partial match as we assume the G has just been identified together with C in the same haplotype and never alone, so the G alone is not known in the database.
