Fastq to Somatic CNV Control
The Fastq to Somatic CNV Control template workflow produces coverage tables that can be used as controls for copy number variant detection.
The workflow can only be used with targeted data.
Use the workflow to generate coverage tables for the Fastq to Annotated Somatic Variants with Coverage Analysis template workflow (Fastq to Annotated Somatic Variants with Coverage Analysis).
Fastq to Somatic CNV Control can be found at:
Template Workflows | LightSpeed Workflows (![]() ) | Fastq to Somatic CNV Control (
) | Fastq to Somatic CNV Control (![]() )
)
If you are connected to a CLC Server via your Workbench, you will be asked where you would like to run the analysis. We recommend that you run the analysis on a CLC Server when possible.
In the first wizard step, select the target regions (figure 4.21).
The target regions must be identical to the target regions that will later be used for copy number variant detection together with the control coverage tables.
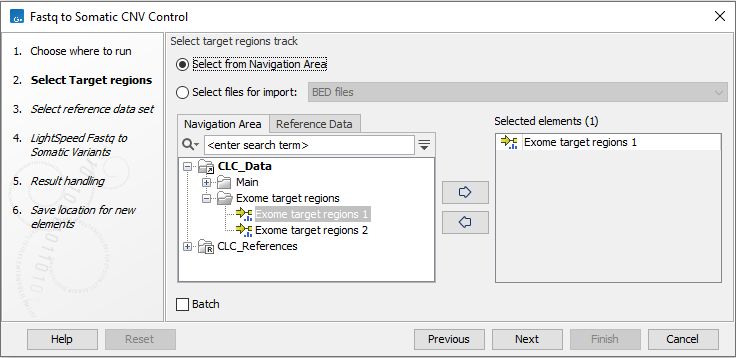
Figure 4.21: Select the target regions.
Next, select a Reference Data Set (figure 4.22). If you have not downloaded the Reference Data Set yet, the dialog will suggest the relevant data set and offer the opportunity to download it using the Download to Workbench button.
If none of the available reference data sets are appropriate, custom reference data sets can be created, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Custom_Sets.html.
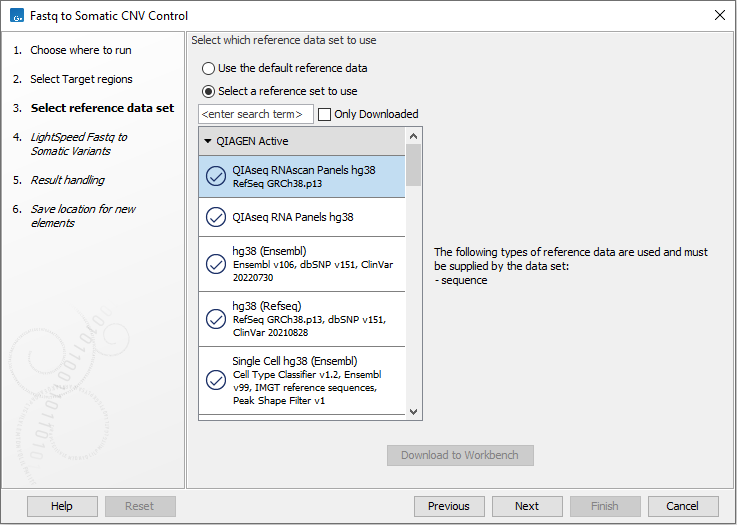
Figure 4.22: Select a reference data set.
In the LightSpeed Fastq to Somatic Variants wizard step (figure 4.23) you have the following options:
- Reads (fastq) Press Browse to select fastq files for analysis.
- Masking mode To enable reference masking when mapping reads, set this option and select a masking track.
- Masking track Provide a masking track for the chosen reference genome if reference masking has been enabled.
- Discard duplicate mapped reads Duplicate mapped reads are per default replaced with a consensus read. Untick if duplicate mapped reads should be retained. See Deduplication for additional details.
- Batch Select if fastq files from different samples are used as input, and each sample should be analyzed individually. The names of the fastq files must follow standard Illumina naming scheme to allow the tool to identify individual fastq files as belonging to the same sample.
- Join lanes when batching Select to join fastq files from the same sample that were sequenced on different lanes.
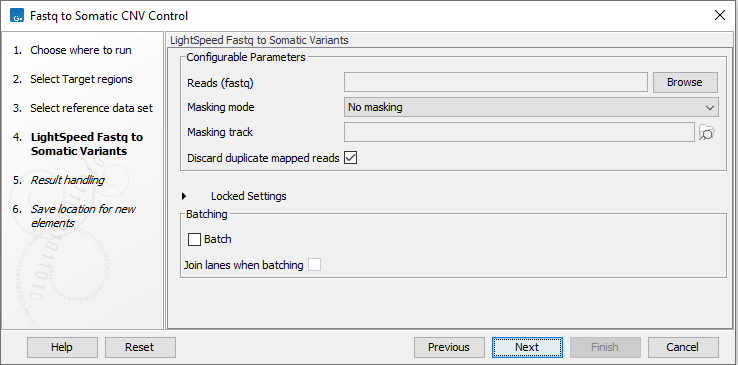
Figure 4.23: Select fastq files.
In the final wizard step, choose to Save the results of the workflow and specify a location in the Navigation Area before clicking Finish.
Subsections
