Launching workflows individually and in batches
When a workflow is launched, a launch wizard appears, allowing you to configure available options. Workflows can be launched by:
- Cicking on the Run Workflow button at the bottom a workflow open in the workflow editor.
-
Selecting an installed workflow from the Workbench Toolbox menu. These are in the following folders:
- Template Workflows This contains workflows included with the software, provided as examples, as described in Template workflows.
- Installed Workflows This contains workflows you have installed in your Workbench. If you are connected to a CLC Server with installed workflows you have access to, those workflows are also listed here.
Launching installed workflows is done the same way as launching tools:
- Select the workflow from the Toolbox menu at the top of the Workbench (as mentioned above).
- Double-click on the workflow name in the listing in the Toolbox tab in the bottom left side of the Workbench.
- Use the Quick Launch (
 ) tool, described in Running tools.
) tool, described in Running tools.
- Template Workflows This contains workflows included with the software, provided as examples, as described in Template workflows.
Workflow inputs
Inputs can be CLC data elements, selected from the Navigation Area, or files located on any accessible system. This includes CLC Server import/export directories when you are connected to a CLC Server, and files on AWS S3 if you have configured an AWS Connection.
To select files from a non-CLC location, choose the "Select files for import" option in the launch wizard (figure 12.43). When you do this, the files will be imported on the fly, as the first action taken when the workflow runs.
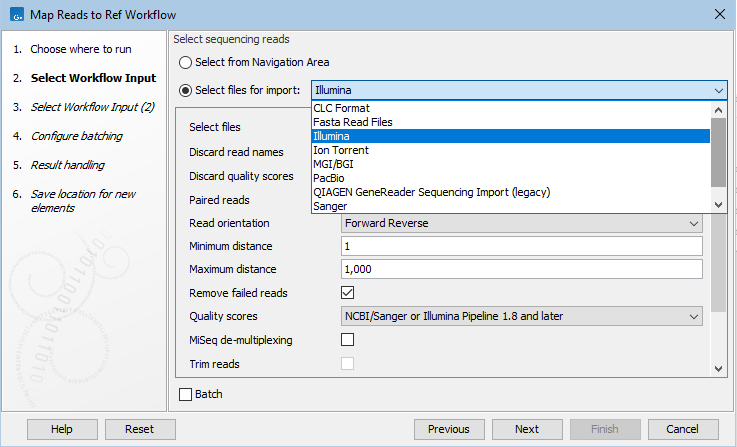
Figure 12.43: To select input data stored somewhere other than a CLC location, choose the option "Select files for import" in the launch wizard. This is often referred to as on-the-fly import.
Further details about workflow inputs are provided in Configuring workflow Input elements. Details about connecting to other systems are in Connections to other systems.
Workflow outputs
Output and Export elements in workflows specify the analysis results to be saved. In addition to analysis results, a Workflow Result Metadata table can be output. This contains a record of the workflow outputs, as described in Workflow Result Metadata tables.
Launching batches of analyses
When multiple inputs are provided, you can choose to run the workflow multiple times, one time for each input, or for defined sets of inputs that should be analyzed together. This is referred to as running in Batch mode, with the inputs to be analyzed together being referred to as batch units. This is described in Running workflows in batch mode.
In addition, using Iterate and Collect and Distribute control flow elements within a workflow allows for part of a workflow being run once per batch unit, while other parts are run on all the data together. This is described in Running part of a workflow multiple times.
Subsections
- Workflow Result Metadata tables
- Running workflows in batch mode
- Running part of a workflow multiple times
