The Upload to QCI Interpret tool
The Upload to QCI Interpret tool can be used to send variant and fusion data, and optionally metadata, to QCI Interpret. It can be found in the Toolbox at:
Tools | QIAseq Panel Expert Tools (![]() ) | QCI Interpret Integration (
) | QCI Interpret Integration (![]() ) | Upload to QCI Interpret (
) | Upload to QCI Interpret (![]() )
)
Select the variant tracks to send to QCI Interpret (figure 14.3), and then select the reference sequence. Hg19 is the default, but hg38 is also supported. You can then choose the analysis type: somatic or hereditary. Note that the somatic and germline/hereditary workflows of QCI Interpret focus on different needs, with somatic being primarily focused on therapeutic, prognostic, and diagnostic actionability, while germline/hereditary is better suited for disease diagnosis/risk.
For RNAscan, Multimodal, or Fusion XP analysis results, specify the relevant fusion tracks for upload in the Fusions section.
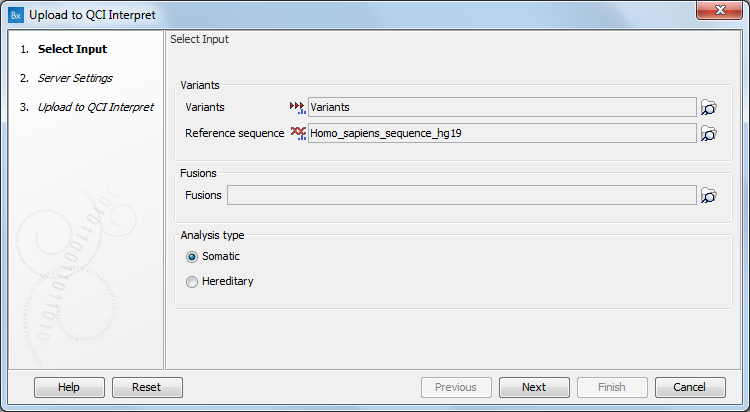
Figure 14.3: The Upload to QCI Interpret dialog.
The next step prompts for the information needed for a successful connection to QCI Interpret, as shown in figure 14.4. Specifically:
- Server location. The specific QCI Interpret server with which your account is associated.
- API key ID and API key secret. See QCI Interpret Integration for information about obtaining these details if you do not already have them.
- Username. Your QCI Interpret username.
For additional information about QCI Interpret user accounts and the QCI Interpret uploader, please refer to the QCI Interpret User Guide, available from the top menu within the QCI Interpret interface.
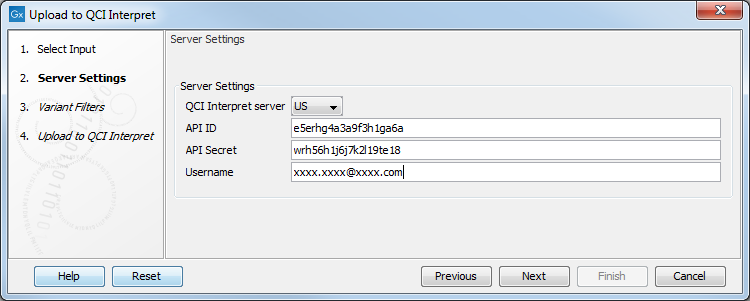
Figure 14.4: Configure the connection to QCI Interpret.
When uploading variant data to QCI Interpret, a dialog called "Variants Filters" will be presented, as shown in figure 14.5. Using these settings, you can limit the list of variants to be uploaded to QCI Interpret to just those of most interest. Restricting the list can be necessary when working with large numbers of variants, particularly with the somatic pipeline, where interpretations are provided for up to 400 variants.
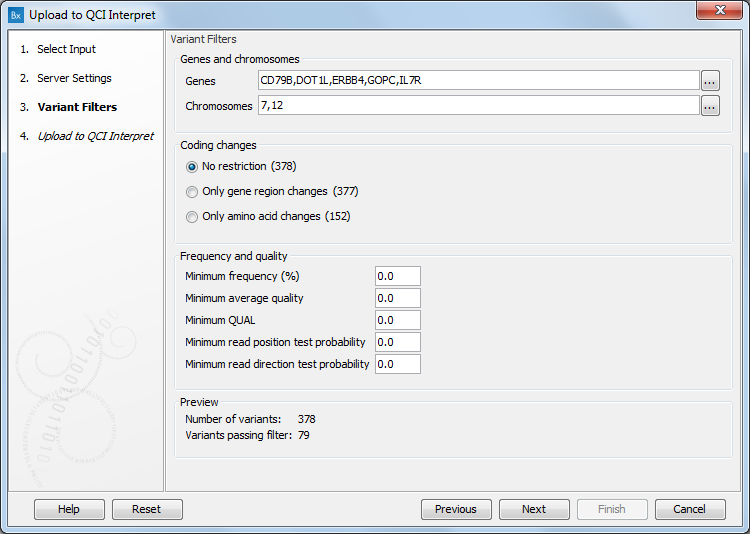
Figure 14.5: The Variants Filters dialog.
You can restrict variants to only those passing filters based on:
- Genes and chromosomes. Click on the buttons to the right of the fields to select the genes or chromosomes of interest.
- Coding changes
- Frequency and quality
The number of variants passing the filters is shown in the Preview section at the bottom of the window, as shown in figure 14.5.
In the last step, click on Finish to initiate the transfer of variants and/or fusions to QCI Interpret. A browser window will open with the QCI Interpret interview page loaded. This page must be filled in and the Submit button pressed to complete the upload of the files and metadata to QCI Interpret. If a browser window fails to open automatically, the tool will output a report containing the link to the QCI Interpret interview page.
