Providing input data for analyses on a CLC Server
All input data must be in a location accessible to the CLC Server8.1. Input data includes the data being analyzed, and in many cases, reference data.
Parameter names for providing inputs reflect the name of the Input element in the workflow design, taking the form <input-element-name>-<parameter-name>.
CLC format data in CLC Server File System Locations or in remote locations accessible via http, https, or S3 URL can be provided as inputs to workflows. Data in other formats or in other locations can be imported at the start of the workflow run using on-the-fly import. These options are described in detail below.
Data in remote locations specified as input for an analysis on the CLC Server is downloaded at the start of the workflow run. Note that AWS charges for data download.
Data can be analyzed directly on AWS, with results saved to AWS S3 using a CLC Genomics Cloud setup, as described in the CLC Cloud Module manual: https://resources.qiagenbioinformatics.com/manuals/clccloudmodule/current/index.php?manual=Overview_CLC_Genomics_Cloud.html.
Providing CLC data as input to workflows
The expected value for each parameter specifying CLC format data to use as input is a ClcServerObjectUrl or an http, https, or S3 URL.
Parameter names for providing CLC format data take the form: <input-element-name>-workflow-input>.
Using on-the-fly import
Data stored somewhere other than a CLC Server File System Location can be supplied as input to analyses by using on-the-fly import. For example, using on-the-fly import, FASTQ sequence files would be imported as the first step in the workflow, avoiding the need for running a specific import command before running the workflow.
On-the-fly import can be used for a particular input if, when a partial workflow command is submitted, the listing of available parameters includes one with a name of the form <input element name>-import-command. The value for that parameter is the importer to use. When on-the-fly import is used, parameters are also needed to specify the location of the files to be imported. One parameter-value pair is needed for each file. The file location is specified using a ClcFileUrl, a ClcCloudFileUrl or an http, https, or S3 URL. An example of using on-the-fly import for import of Illumina data is provided below.
Many importers require additional configuration information. These options parameter are listed when the incomplete clcserver command includes the <input element name>-<import-command> parameter. The settings for individual importers are described in the import chapter of the CLC Genomics Workbench manual: https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Importing_data.html.
On-the-fly import is usually possible when an Input element has been connected to the relevant input channel in the workflow design. However, a workflow author can configure Input elements so on-the-fly import is not allowed (see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Configuring_Workflow_Input_elements.html).
Specifying input data - an example
The parameters in this section relate to the workflow shown in figure 8.1. There are 2 Input elements in this workflow, Sample Reads and Reference Genome.
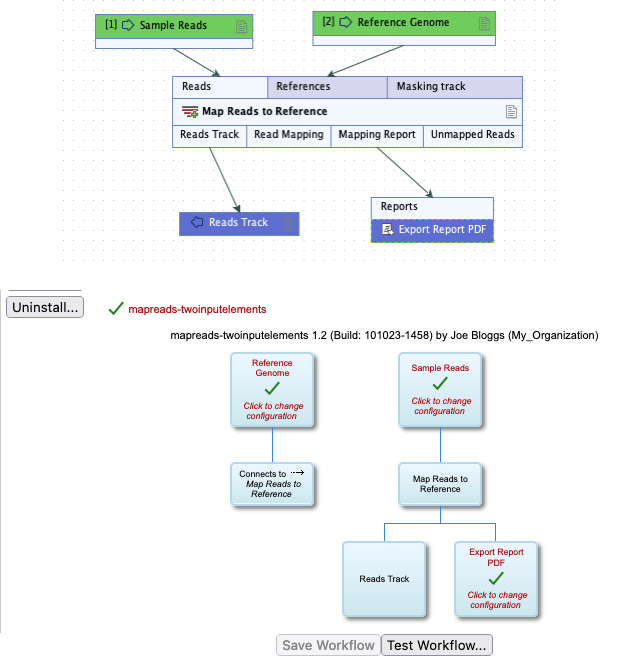
Figure 8.1: A simple workflow seen in the Workflow Editor of a CLC Workbench (top) and the CLC Server web interface (bottom). Two Input elements are present, Sample Reads and Reference Genome.
Using this workflow, a command of the following form:
clcserver -S <server> -U <username> -W <password or token> -A wf-mapreads-twoinputelements
would return the following parameter information relating to inputs:
--reference-genome-import-command < The importer to use for on-the-fly import of input data. [clc_import, ngs_import_fasta, ngs_import_genereader, ngs_import_illumina, ngs_import_iontorrent, ngs_import_mgi_bgi, ngs_import_pacbio, ngs_import_sanger, trace_files_import]> --reference-genome-workflow-input Workflow Input <ClcObjectUrl> --sample-reads-import-command < The importer to use for on-the-fly import of input data. [clc_import, ngs_import_fasta, ngs_import_genereader, ngs_import_illumina, ngs_import_iontorrent, ngs_import_mgi_bgi, ngs_import_pacbio, ngs_import_sanger, trace_files_import]> --sample-reads-workflow-input Workflow Input <ClcObjectUrl>
For the Sample Reads input element, the parameters available are:
--sample-reads-workflow-inputUsed to specify CLC format data in a CLC Server File System Location, with a CLC Object URL as the value, or in a remote location, with an http, https, or S3 URL as the value.One parameter-value pair is needed for each input element.--sample-reads-import-commandUsed to indicate that the data should be imported as the first step of the workflow (on-the-fly import). One of the available importers is expected as the value. Corresponding--sample-reads-select-filesparameters, each with a ClcFileUrl as the value, are then used to specify each file to import. One parameter-value pair is needed for each input file.
All data for a given workflow input (e.g. Sample Reads) must either be specified using
--sample-reads-workflow-input parameters or it must be imported using
a --sample-reads-import-command parameter and --sample-reads-select-files parameters.
A common situation is to use on-the-fly import for sample data and to use already imported data for reference data. Doing this for the example workflow, the input related parameters could look like:
--reference-genome-workflow-input clc://server/<path-to>/<reference-data-element> \ --sample-reads-import-command ngs_import_illumina --reads-paired-reads false \ --sample-reads-select-file clc://serverfile/<path-to>/SRR6954665_R1.fastq \ --sample-reads-select-file clc://serverfile//<path-to>/SRR6954668_R1.fastq \ --sample-reads-select-file clc://serverfile//<path-to>/SRR6954672_R1.fastq \ --sample-reads-select-file clc://serverfile//<path-to>/SRR6954680_R1.fastq \ --sample-reads-select-file clc://serverfile//<path-to>/SRR6954681_R1.fastq \ --sample-reads-select-file clc://serverfile//<path-to>/SRR6954682_R1.fastq
Obtaining QIAGEN reference data
Many template workflows, and copies of such workflows, refer to reference data supplied by QIAGEN. A CLC Genomics Workbench is needed to download these data elements. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=QIAGEN_Sets.html for information on how to do this. This data must be downloaded to the CLC Server, which must have a CLC_References location configured. For further details, see https://resources.qiagenbioinformatics.com/manuals/clcserver/current/admin/index.php?manual=Reference_data_management.html.
Footnotes
- ... Server8.1
- The information in this section relates to launching analyses to run on the CLC Server. Please refer to the CLC Cloud Module manual for information relevant to submitting analyses to run on a CLC Genomics Cloud setup.
