References management
The ready-to-use workflows rely on the presence of particular reference data sets. The References management tool (figure 8.1) in the workbench makes it easy to download the necessary data ahead of time, and to create custom sets when needed.
The References management tool (figure 8.1) offers an easy way of retrieving popular reference data sources such as genes, variant annotations and genome sequences as tracks.
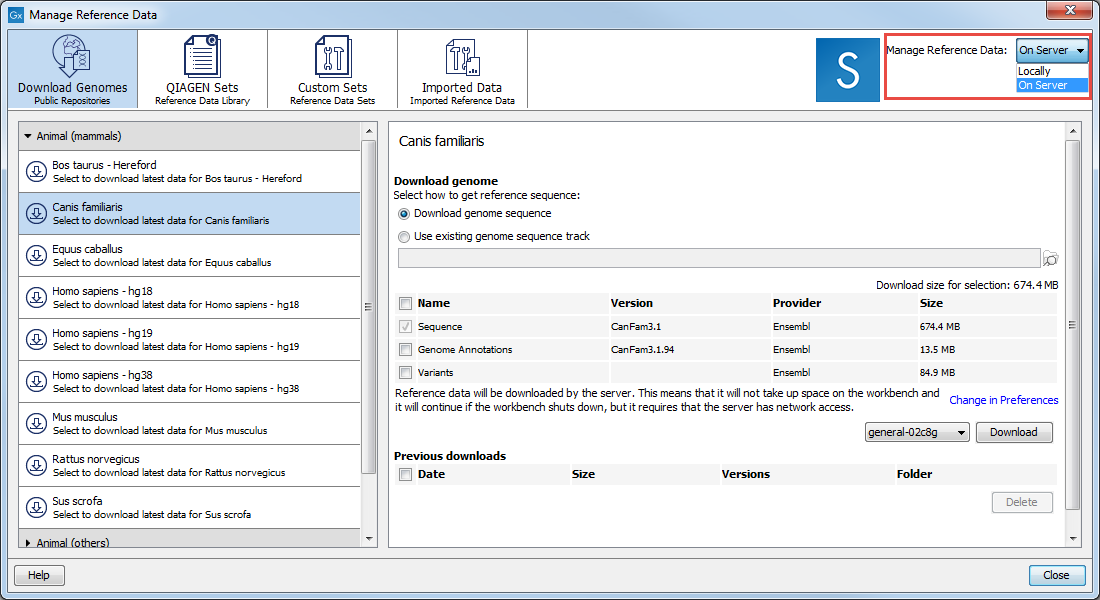
Figure 8.1: Click on the References button and choose the QIAGEN Sets tab to find and download reference data; Custom Sets to customize reference data sets; and Imported Data to import your own reference data.
The total size of the reference data you can download can vary and is indicated when selecting the elements to download. The amount of time it will take to download this data depends on your network connection, but it can take several hours on slower connections.
Where reference data is downloaded from
Data download using the Download Genomes comes from public repositories such as Ensembl, NCBI, UCSC. This type of data is not provided or hosted by QIAGEN. The workbench only provides an easy way to retrieve data that should otherwise have been downloaded and imported. The list of organisms is dynamically updated by QIAGEN independently of Workbench versions, so you will always see the most recent list of organisms. If you do not find the organism you are looking for, there is always the possibility to download and import the data using the Import tools.
The QIAGEN Sets tab allows you to download curated Reference Data Sets directly from a QIAGEN reference data repository. The location of the repository can be changed in the Edit | Preferences | Advanced as shown in figure 8.2.
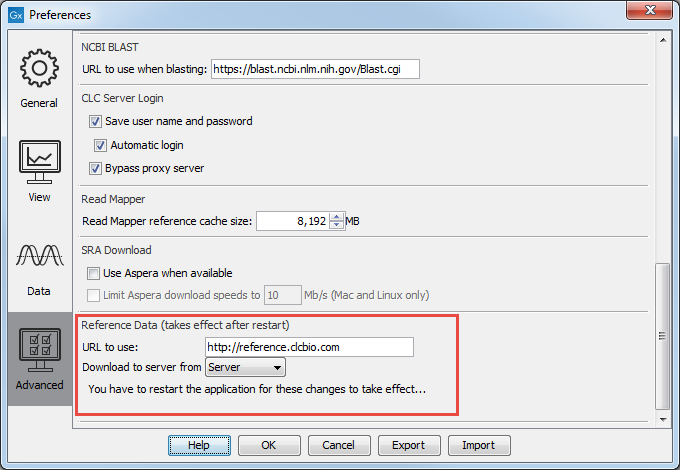
Figure 8.2: The location where reference data is downloaded from can be seen in the Workbench Preferences.
Unless you are in the special circumstance that your system administrator has decided to mirror this data locally and wishes you to use that mirror of the data, you should not change this setting.
Where reference data is downloaded to
The reference data that is downloaded will be stored in a folder called CLC_References. When the CLC Genomics Workbench is installed, such a folder is created on your file system under your home area. This folder is specified within the workbench as a reference location.
On the right hand side of the Reference manager, you can use the drop-down menu to choose where you intend to manage the reference data. If you choose "Locally", the Download and Delete buttons will work on the local reference data. If you choose "On Server" (only available if you are connected to a server), the buttons will work on the reference data on the server you are connected to (figure 8.3).
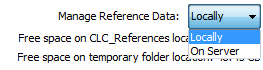
Figure 8.3: Reference data can be available locally or on the server.
When working locally, you can also check how much free space is available for the Reference folder on your local disk.
When working on a Server, a drop down menu situated below the list of existing element to download for a particular species in the Download Genomes tab will let you choose which server or grid node to use for the download.
When the option "On Server" is selected and you are working with a server configured with grid nodes, then a drop down menu listing the available grid presets will be present to the left of the Download button when working with data available under the Download Genomes tab. Select the correct grid before pressing the Download button.
Changing the reference data location
You can specify a different location to download reference data to. This is recommended if you do not have enough space in the area the workbench designates as the reference data location by default. To change the reference data location from within the Navigation Area:
Right-click on the folder "CLC_References" | Choose "Location" | Choose "Specify Reference Location"
The new folder will also be called CLC_References, but will be located where you specify.
In more detail, this action results in the following:
- A folder called CLC_References is created in the location you specified, if a folder of this name did not already exist.
- The workbench sets this new location as the place to download reference data to and the place the ready-to-use workflows should look for reference data.
This action does not:
- Remove the old CLC_References folder.
- Remove the contents of the old CLC_References folder, such as previously downloaded data.
If you have previously downloaded data into the CLC_References folder with the old location, you will need to use standard system tools to delete this folder and/or its contents. If you would like to keep the reference data from the old location, you can move it, using standard system tools, into the new CLC_References folder that you just specified. This would save you needing to download it again.
Note! If you run out of space, and realize that the CLC_References should be stored somewhere else, you can do this by choosing a new location, then manually moving the already downloaded files to that new location, and restarting the workbench. The "downloaded references" file will then be updated with all the new references.
Reference data for non-networked systems
CLC Genomics Workbench may be installed on computers that have no access to the external network. In that case, please proceed with the following steps to import reference data to the non-networked workbench:
- Install CLC Genomics Workbench on a machine with access to the external network.
- Download an evaluation license via the Workbench License Manager. If you have problems obtaining an evaluation license this way, please write to us at ts-bioinformatics@qiagen.com.
- Use the Reference Data Manager on the networked Workbench to download the reference data of interest. By default, this would be downloaded to a folder called CLC_References.
- When the download is completed, copy the CLC_References folder and all its contents to a location where the machines with the CLC software installed can access it.
- Get the software to refer to that folder for reference data: in the Navigation Area of the non-networked Workbench, right click on the CLC_References, and choose the option "Specify Reference Location...". Choose the folder you imported from the networked Workbench and click Select.
You can then access reference data using the Reference Data manager.
Subsections
