Running the Transcript Discovery tool
To run the Transcript Discovery tool, go to:
Tools | RNA-Seq Analysis (![]() ) | Transcript Discovery (
) | Transcript Discovery (![]() ) | Transcript Discovery
) | Transcript Discovery
Select the read mapping produced by the Large Gap Read Mapping tool (figure 3.1).
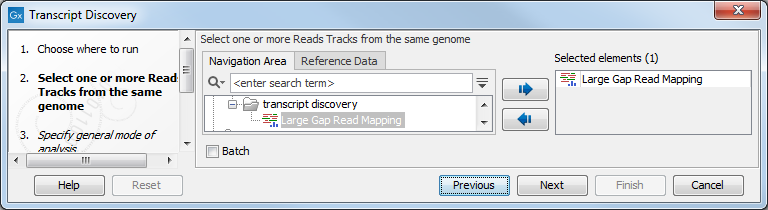
Figure 3.1: Specify a Large Gap Read Mapping track.
You are now presented with choices regarding the overall mode of analysis as shown in figure 3.2.
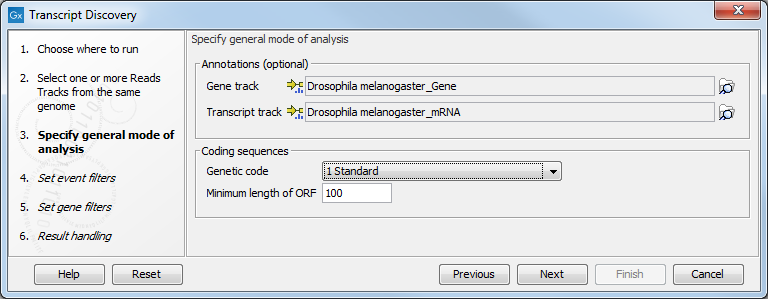
Figure 3.2: Specifying the overall mode of analysis.
You can specify a gene and a transcript track to add annotations to your analysis, but this is optional. You can also choose the Genetic code from a drop down menu, and the Minimum length of ORF desired.
In the Set event filters dialog figure 3.3, the options are as follow:
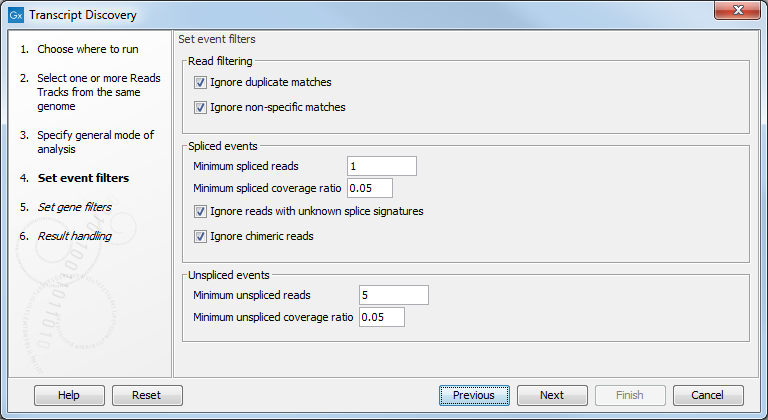
Figure 3.3: Specifying the event filters.
- Read filtering
- Ignore match duplicates. For reads that are 100% identical, only one copy is used to define events. This is relevant for the 'supporting read counts' that are used when filtering events. When ticked, identical reads will only be counted as '1' in the read counts.
- Ignore non-specific matches. Reads that have an equally good match elsewhere on the reference genome (these reads are colored yellow in the mapping view) can be ignored in the analysis. Whether you include these reads or not will be a tradeoff between sensitivity and specificity. Including them may lead to the prediction of transcripts that are not correct, whereas excluding them may mean that you will lose some true transcripts.
- Spliced events
- Minimum spliced reads. This filter removes spliced events with weak evidence by defining the minimum number of unique spliced reads that must support a spliced event. Events that do not meet this requirement are ignored. A read is unique if it 'counts' as specified by the 'Read filtering' options: if the 'Ignore duplicate reads' option is checked, identical spliced reads are counted as 1, and if the 'Ignore non-specific matches' is checked, non-specific matches are not counted. Minimum spliced reads is set by default to 1, meaning that by default no filtering is happening.
- Minimum spliced coverage ratio. This filter removes spliced events with weak relative evidence. The spliced coverage of a region is calculated as the number of spliced reads in the gene region, divided by the total length of the region consisting of the union of the exons in the events in the region. Similarly, the spliced coverage of an event is calculated as the number of spliced reads supporting the event, divided by the length of the exon regions of the event. If the spliced coverage of the event divided by the spliced coverage of the region is smaller than this value, the event is ignored. Compared to the filter on absolute read count above, the coverage ratio filter allows filtering of events with weak evidence in regions of high coverage.
- Ignore reads with unknown splice signatures. As canonical splice signatures are overwhelmingly found (GT - AG, GC - AG and AT - AC), this filter typically removes badly mapping reads that do not fit theses signatures and that are likely assembly errors. Consequently, this filter removes events containing uncertain positions (see figure 3.4 for an example of how such an event is represented in the Workbench).

Figure 3.4: Uncertain position as depicted in a track view. - Ignore chimeric reads. This filter removes spliced events that span two or more genes. Keeping these events will typically cause us to merge the genes in the output. The filter works by assigning spliced events to possible "chains" of exons. Events are chained if they are within 1000bp of each other. This value is hard-coded, and is chosen as a good upper-bound on exon length. Note that this length is unrelated to the "Gene merging distance" parameter, because that parameter looks for gaps in coverage between any events, whereas this length only cares about distances between spliced events. If a spliced read passes over a chain end and then a chain start, in that order, and on the same strand as itself, then it is inferred to have spanned two genes.
- Minimum spliced reads. This filter removes spliced events with weak evidence by defining the minimum number of unique spliced reads that must support a spliced event. Events that do not meet this requirement are ignored. A read is unique if it 'counts' as specified by the 'Read filtering' options: if the 'Ignore duplicate reads' option is checked, identical spliced reads are counted as 1, and if the 'Ignore non-specific matches' is checked, non-specific matches are not counted. Minimum spliced reads is set by default to 1, meaning that by default no filtering is happening.
- Unspliced events
- Minimum unspliced reads. This filter removes events with weak evidence by defining the minimum number of unique unspliced reads that must support an unspliced event. Events that do not meet this requirement are ignored. It is set by default to 5.
- Minimum unspliced coverage ratio. This filter removes events with weak relative evidence. The un-spliced coverage of a region is calculated as the number of un-spliced reads in the transcript event region, divided by the total length of the region consisting of the union of the exons in the events in the region. Similarly, the un-spliced coverage of an event is calculated as the number of un-spliced reads supporting the event, divided by the length of the exon regions of the event. If the un-spliced coverage of the event divided by the un-spliced coverage of the region is smaller than this value, the event is ignored.
In the Set gene filters dialog figure 3.5, the options are as follow:
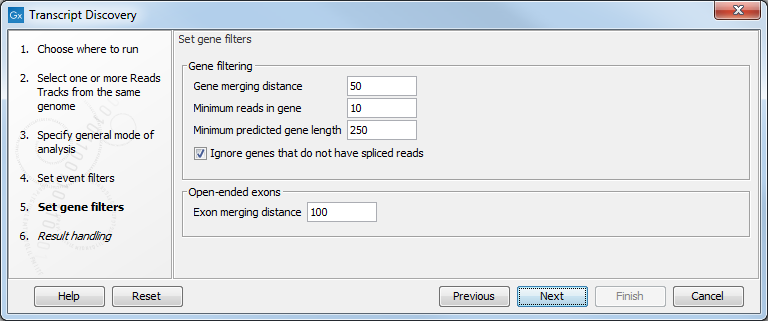
Figure 3.5: Specifying the gene filters.
- Gene filtering
- Gene merging distance. If two reads map closer than this value and on the same strand, then they are considered to be part of the same gene. This closeness criterion is re-evaluated several times during the process. For example, if an event is filtered away, it may create a gap greater than this size, leading to a new gene region. Setting this value too high may cause genes to be merged together. Setting the value too low may cause genes to be split in two or more pieces in low coverage regions. Genes supplied as known annotations are not merged.
- Minimum reads in gene. This filter will remove predicted genes that have fewer supporting reads than this number. Note that genes supplied as known annotations are not ignored. This parameter is set at 10, which is suitable for Illumina reads, but can be lowered to 2 when working with very long reads such as PacBio data.
- Minimum predicted gene length. This filter will remove genes predicted to be shorter than this value. Note that genes supplied as known annotations are not ignored.
- Ignore genes that do not have spliced reads. This filter will remove genes that do not have spliced reads. Note that genes supplied as known annotations are not ignored. This option, on by default, can be turned off when working with PacBio long reads.
- Open-ended exons
- Exon merging distance. Open ended exons that are within this distance of each other, and which do not have a splice junction between them will be joined together into one exon. This helps reconstruct full-length exons from regions with low coverage. Exons supplied as known annotations are not merged.
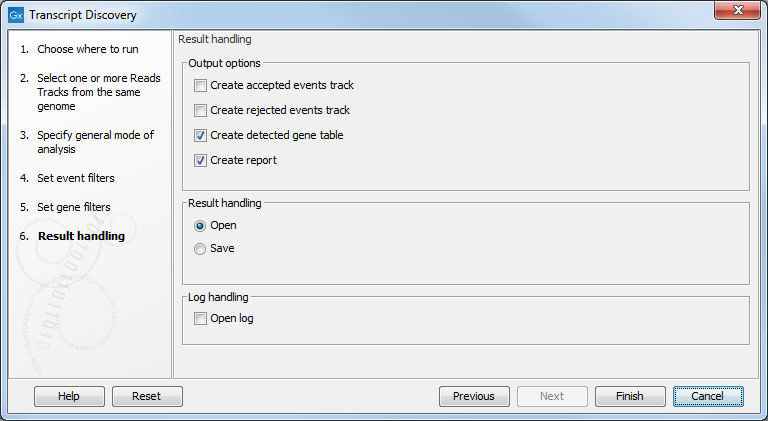
The Transcript Discovery tool can generate the following outputs (see 3.6).