Tabular mapping files
The CLC Genomics Workbench supports import and export of files in tabular format such as Eland files coming from the Illumina Pipeline. The importer is quite flexible which means that it can be used to import any kind of mapping file in a tab-delimited format where each line in the file represents one read.
The idea behind the importer is that you import the mapping file which includes all the reads and then you specify one or more reference sequences which have already been imported into the Workbench. The Workbench will then combine the two to create mapping results (![]() ) or mapping tables (
) or mapping tables (![]() ). To import a tabular mapping file:
). To import a tabular mapping file:
File | Import High-Throughput Sequencing Data (![]() ) | Tabular Mapping Files (
) | Tabular Mapping Files (![]() )
)
This will open a dialog where you choose the reference sequences to be used as shown in figure 6.16.
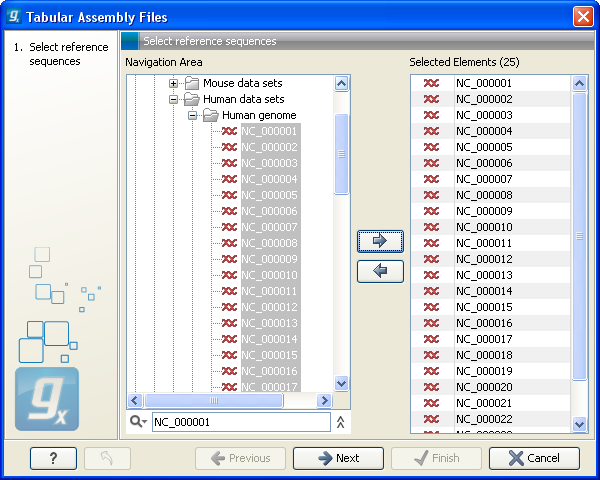
Figure 6.16: Defining reference sequences.
Select one or more reference sequence. Note that the name of your reference sequence has to match the reference name specified in the file. Click Next.
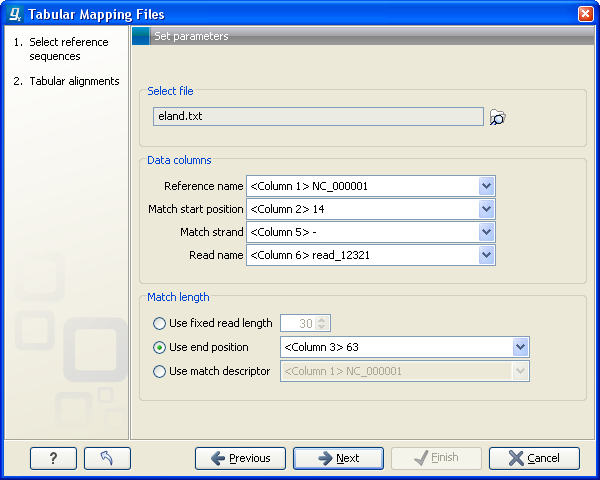
Figure 6.17: Defining reference sequences.
In this dialog, select (![]() ) one or more tab delimited files as shown in figure 6.17.
) one or more tab delimited files as shown in figure 6.17.
Once the tab delimited file has been selected, you have to specify the following information:
- Data columns. The Workbench needs to know how the file is organized in order to create a result where the reads have been mapped correctly.
- Reference name. Select the column where the name reference sequence is specified. In the example above, this is in column 1.
- Match start position. The position on the reference sequence where the read is mapped. The numbering starts from position 1.
- Match strand. Whether the read is mapped the positive or negative strand. This should be specified using
F/R(denoting forward and reverse reads) or+/-. - Read name. Whether the read is mapped the positive or negative strand. This should be specified using
F/R(denoting forward and reverse reads) or+/-.
- Match length. The start position of the read is set above. In this section you specify the length of the match which can be done in any of the following ways:
- Use fixed read length. If all reads have the same length, and if the read length or match end position is not provided in the file, you can specify a fixed length for all the reads.
- Use end position. If you have a match end position just as a match start position, this can be used to determine match length.
- Use match descriptor. This can be used to denote mismatches in the alignment. For a 35 base read,
35denotes an exact match and32C2denotes substitution of aCat the 33rd position.
Click Next to adjust how to handle the results. We recommend choosing Save in order to save the results directly to a folder, since you probably want to save anyway before proceeding with your analysis.
Note that this import operation is very memory-consuming for large data sets.
