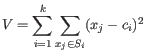K-medoids clustering is computed using the PAM-algorithm (PAM is short for Partitioning Around Medoids). The PAM-algorithm is based on the search for
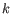
representatives (called medoids) among all elements of the dataset. When having found
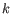
representatives,
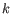
clusters are generated by assigning each element to its nearest medoid. The algorithm first looks for a good initial set of medoids (the BUILD phase). Then it finds a local minimum for the objective function:
where there are
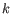
clusters
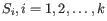
and
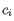
is the medoid of
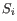
. This solution implies that there is no single switch of an object with a medoid that will decrease the objective (this is called the SWAP phase). The PAM-agorithm is described in [
Kaufman and Rousseeuw, 1990].
Features are z-score normalized prior to clustering: they are rescaled such that the mean expression value over all input samples for the clustering is 0, and the standard deviation is 1.