Annotate Variants with Unique Molecular Index Info
The tool Annotate Variants with Unique Molecular Index Info annotates the variants with UMI groups information generated by the Calculate Unique Molecular Index Groups, and produces the annotated variant track as output.
The tool can be found in the Toolbox here:
Tools | QIAseq Panel Expert Tools | QIAseq DNA Panel Expert Tools (![]() ) | Annotate Variants with Unique Molecular Index Info (
) | Annotate Variants with Unique Molecular Index Info (![]() )
)
In the first dialog (figure 3.47), select a variant track.
Figure 3.47: Select a variant track.
In the second dialog, select a read mapping. The tool works on any read mapping on which UMI groups have been calculated, i.e. a read mapping consisting of raw reads or a read mapping consisting of UMI consensus reads generated by the Create UMI Reads tool (as seen on figure 3.48). If the read mapping consists of UMI reads, check the "Mapping consists of UMI reads" option.
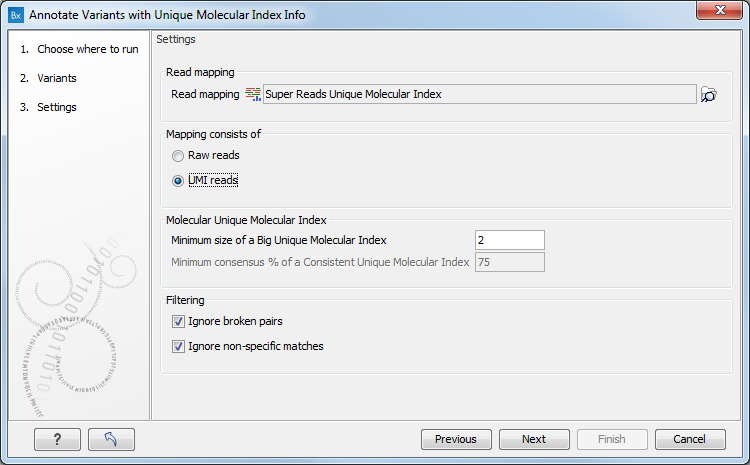
Figure 3.48: Select a read mapping.
The parameters below are used to calculate the annotations:
- Minimum size of a Big Unique Molecular Index: Minimum number of reads in a UMI group for it to be considered Big.
- Minimum consensus % of a Consistent Unique Molecular Index: Minimum percentage of reads in a UMI group that should support a variant for the UMI to be considered Consistent for that variant. This option is valid only if the read mapping chosen is made of raw reads.
Finally, it is possible to filter the data using the following options:
- Ignore broken pairs: reads from broken pairs will be ignored.
- Ignore non-specific matches: read that map in multiple places will be ignored.
Annotations
The following annotations are added to the variants found using a read mapping consisting of raw reads, while only the three annotations indicated with a * are added when the read mapping consists of UMI reads. When using the Analyze QIAseq DNA Panels guide or the Identify QIAseq DNA Variants workflow, the annotations are always based on UMI reads.
- Coverage (UMI): Number of UMI groups that overlap this variant. It is the coverage in the UMI reads track as seen by the Annotate Variants with Unique Molecular Index Info tool. Note that this value can be different form the Coverage value, which is based on the coverage in the UMI reads track as seen by the Low Frequency Variant Detection tool, where broken pairs, non-specific reads and reads with pyro-error variants are filtered out when using the default settings.
- Coverage (Big UMI): Number of big UMI groups that overlap this variant.
- Count (UMI): Number of UMI groups where at least one read has this variant.
- *Count (singleton UMIs): Number of singletons UMIs supporting the variant.
- *Count (big UMIs): Number of big UMIs supporting the variant.
- Count (Consistent and Big UMI): Number of Consistent and Big UMI groups that have this variant.
- *Proportion (singleton UMIs): Proportion of UMIs supporting the variant that are singleton UMIs.
- Freq (UMI): The percentage of UMI groups with this variant out of all UMI groups overlapping this variant.
- Freq (Consistent and Big UMI): The percentage of Consistent and Big UMI groups out of all UMI groups overlapping this variant.
- F/R (UMI coverage): Forward reverse balance of the UMI groups that overlap this variant.
- F/R (UMI count): Forward reverse balance of the UMI groups that have this variant.
- F/R (Big UMI coverage): Forward reverse balance of the Big UMI groups that overlap this variant.
- F/R (Consistent and Big UMI count): Forward reverse balance of the Big and Consistent UMI groups that have this variant.
- UMI info: A value of "24/29; 6/8; 1/40 (12 total)" means that there are 12 UMI groups with at least 1 read having this variant, the best of these groups consist of 29 read, where 24 of those reads have this variant, the second best group have 6 our of 8 reads with this variant. A variant can be overlapped by paired read that overlaps itself, where only the left or the right end has the variant. As long as at least one of the left or right ends of the paired read has the variant, we count the paired read as having the variant.
The annotations differ when the tool is used with UMI reads as it is not possible to calculate annotations involving how many reads in a read group have the variant from the UMI consensus reads. This is for example "Found matching groups with good part matching", "Found matching groups by reads and group sizes", etc.. These columns then assume that all reads in a read group have the variant, that means that all matching UMI groups are Consistent. It is also not possible to calculate how many reads match a variant and how many do not. Many columns show a frequency of reads and also a frequency of groups, e.g. "Found matches" and "Found matches UMI-groups". When running on UMI reads, these two numbers will be the same.
