Download Microbial Reference Database
The Download Microbial Reference Database tool downloads selected references from GenBank, and outputs a single sequence list with the necessary annotations required for the tools in the Typing and Epidemiology and Metagenomics sections of the Microbial Genomics Module.
To run the tool, go to:
Databases (![]() ) | Taxonomic Analyses (
) | Taxonomic Analyses (![]() ) | Download Microbial Reference Database (
) | Download Microbial Reference Database (![]() )
)
In the first window (figure 20.1), select the source of the database you wish to generate.
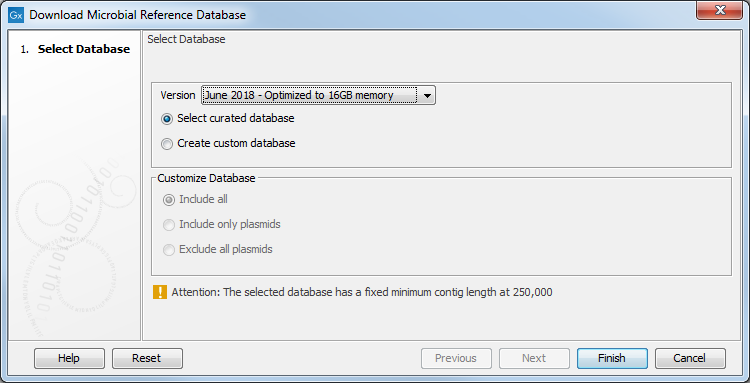
Figure 20.1: Select the references you want to download.
By choosing Select curated database, you can choose to download a database which is optimized for balance in the taxonomic representation across the taxonomy, i.e. the oversampling of some branches of the taxonomy is removed by using representative sequences. This has the consequence that some assemblies may not be particularly good assemblies, yet they are included as they constitute the best current representative of the given branch in the taxonomy. For this optimized database you can choose to download the full database, or one that is optimized for running the Taxonomic Profiling tool on a laptop computer with 16GB of main memory. The two versions of the curated database contain the same assemblies, but the database that is adapted for running on a system with 16GB of main memory does not contain contig sequences shorter than 250,000 bp.
If you select Create custom reference database, then the section Customize Database will become active. Select one of the three options:
- Include all: The database will contain both genomic and plasmid sequences.
- Include only plasmids: The database will contain only plasmid sequences.
- Exclude all plasmids: The database will not contain any plasmid sequences.
Click Next to select the source of the database you wish to generate (as in figure 20.2). Here you can provide a list of GenBank or RefSeq assembly accessions that must be included in your database. The assessions must follow the assembly accession: 3 letter prefix, (GCA for GenBank assemblies or GCF for RefSeq assemblies) followed by an underscore and 9 digits. For example, GCA_000019425 for the assembly of the DH10B substrain of E. coli. If a version number is included, it will be ignored and the newest version downloaded.
The GenBank assembly accession can usually be accessed using the search function or from the GenBank sequence on NCBI.
You have the following options:
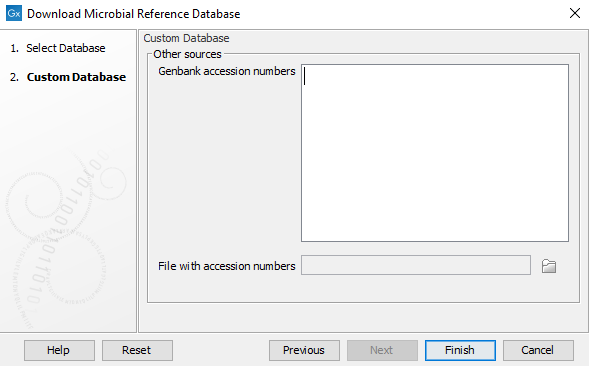
Figure 20.3: Optional: Input accession numbers to include in the database download
- Provide a list of Genbank accession numbers in the white field, or
- Browse your computer for a file with accession numbers, or
- Leave fields blank and press Finish: the database will not have any references pre-selected. Selection can be done manually.
The required metadata for building a selection table will now be downloaded. No assembly data is downloaded at this point. This process will take a brief moment.
The tool will open a table called a Database builder (figure 20.3) from which you can design your own database. A series of functionality can help you filter and sort the table to extract the information relevant to your project.
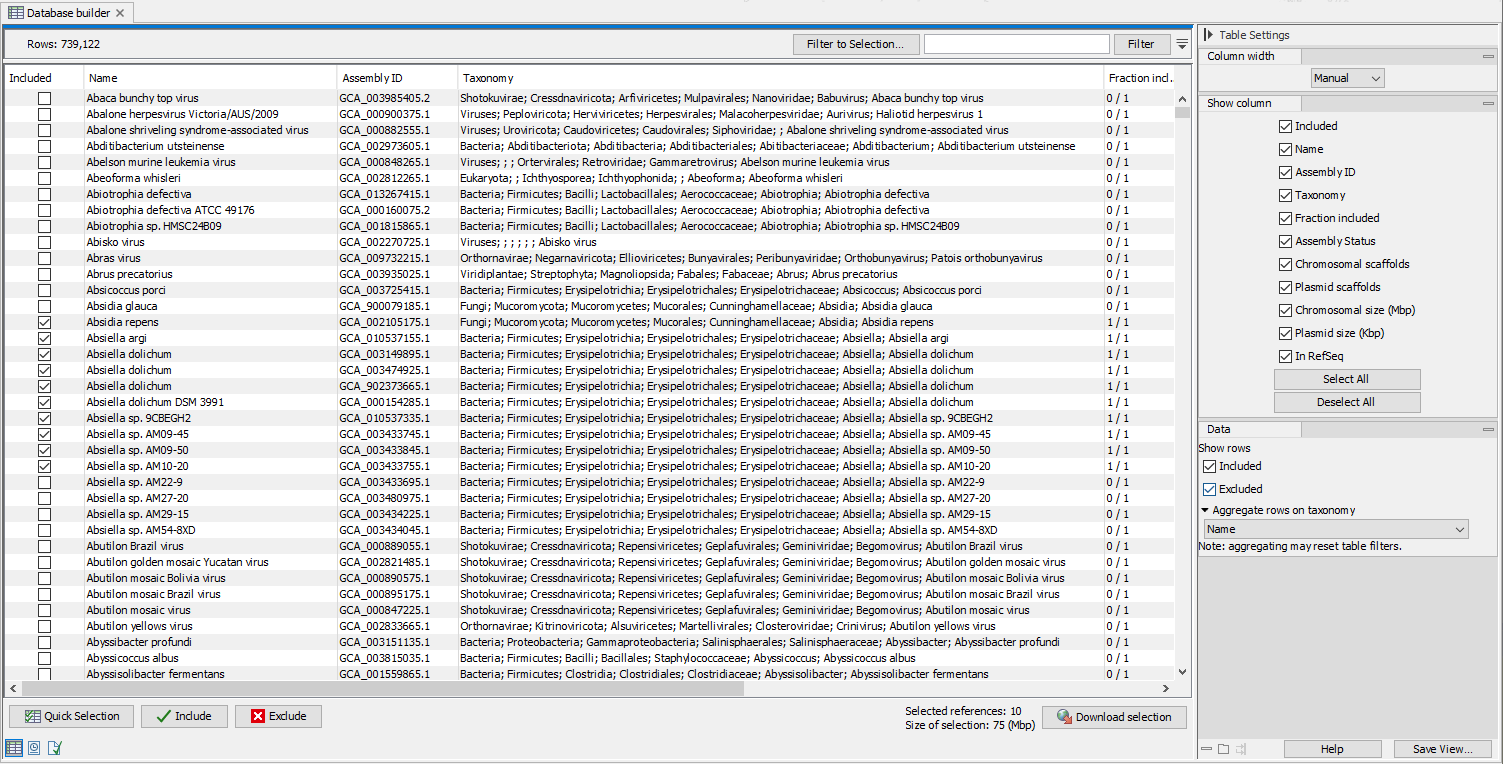
Figure 20.4: Search, filter and select assemblies to download
- Use the "Quick selection" button to quickly select predefined subsets for
download:
- Single scaffold complete genomes in RefSeq
- Complete genomes in RefSeq
- All complete genomes
- Aggregate the table to a specified taxonomic group using the drop down menu in the "Data" palette of the side panel. Use the category "Name" to de-aggregate the table.
- Use filter(s) and select row by dragging or pressing Ctrl+A to keep only the rows you are interested in, and click on the button Include to stage the selected references for downloading, which is indicated by a checkmark in the "Included" column.
- Alternatively, press Ctrl+A then click on Include to include all rows first. Then set one or several filters, and use the button Exclude on the remaining rows. Clear the filter(s) by clicking on the red buttons next to each filter set. The rows not filtered away in the second step should still be checked.
Once the table has all the desired references included, which is indicated by a checkmark, click Download selection. Close to the button, you can check how many references are selected and see an estimate of the total size of the selection.
The dialog shown in figure 20.4 allows you to include all annotation tracks (annotation tracks are not needed for taxonomic profiling applications, but required when creating Large MLST schemes). An additional filter, Minimum contig length, may also be specified (this option is not available when downloading the curated database). It also warns about the memory and disk requirements that will be needed to later run the Taxonomic Profiling tool with the database you are about to download.
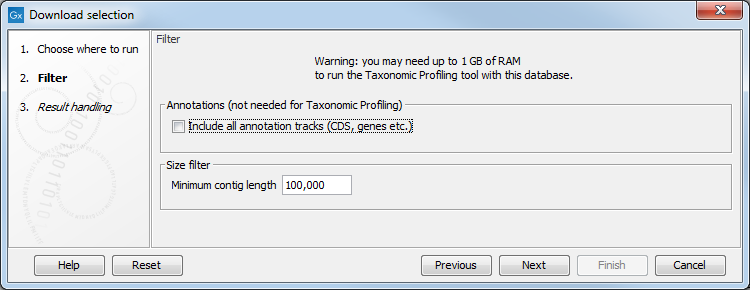
Figure 20.2: The Download selection wizard.
