Mapping output options
Click Next lets you choose how the output of the mapping should be reported (see figure 25.9).
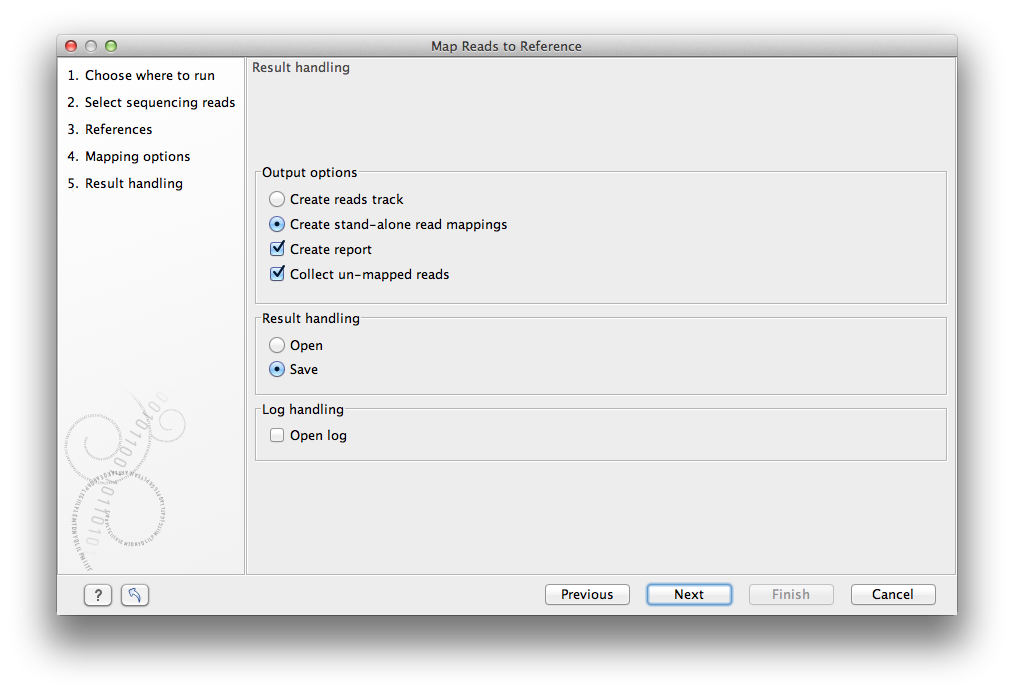
Figure 25.9: Mapping output options.
The main choice in output format is at the top of the dialog - the read mapping can either be stored as a track or as a
stand-alone read mapping. Both options have distinct features and advantages:
- Reads track
- A reads track is very "lean" (i.e. with respect to memory requirements) since it only contains the reads themselves. Additional information about the reference, consensus sequence or annotations can be added and viewed alongside in the context of a Track List later (by adding, for example, a reference and/or annotation track, respectively). This kind of output is useful when working with tracks in general and especially for resequencing purposes this is recommended. Details about viewing and editing of reads-tracks are described in tracks, and the resequencing.
The main advantage of having the output of the read-mapping process represented as a track is that this way it seamlessly integrates with other downstream analysis tools. In contrast, the stand-alone read mapping output has a couple of specialized functions which are not directly available for single reads-tracks but requires the context of the overall tracks-framework. However, unless any specific functionality of the stand-alone read-mapping is required, we recommend to use the tracks output for the additional flexibility in further analysis. Later it is possible to convert to and from tracks (see Converting data to tracks and back). The side-panel functionality for viewing a read track and an annotation track is shown in Figure 24.7.
- Stand-alone read mapping
- This output is more elaborate than the reads track and includes the full reference sequence (including annotations) and a consensus sequence is created as part of the output. Furthermore, the possibilities for detailed visualization and editing are richer than for the reads track (see View and edit read mapping). The down-side of a stand-alone read mapping is that it copies all the information from the reference sequence which can take up a lot of disk space, and second that it does not lend itself to comparative analyses. If you wish to compare e.g. SNPs from one sample to another sample, or against a database of variants, this calls for using a reads-track instead. Note that if multiple reference sequences are used as input, a read mapping table is created (see Mapping table).
In addition to the choice between the two main output options, there are two independent output options available that can be (de-)activated in both cases:
- Create report. This will generate a summary report as described in Summary mapping report.
- Collect un-mapped reads. This will collect all the reads that could not be mapped to the reference into a sequence list (there will be one list of unmapped reads per sample, and for paired reads, there will be one list for intact pairs and one for single reads where the mate could be mapped).
Finally, you can choose to save or open the results, and if you wish to see a log of the process (see how to handle the results).
Clicking Finish will start the mapping.

