Output of the Differential Expression tools
Statistical comparison tracks
The Differential Expression for RNA-Seq tool will output one or more statistical comparison tracks or tables. The statistical comparison table offers the same functionality than the track, except for the track view.
An example of a statistical comparison track is shown in figure 31.80. Statistical comparison tracks make it possible to show differential expression data alongside other kinds of tracks in a genomic context.
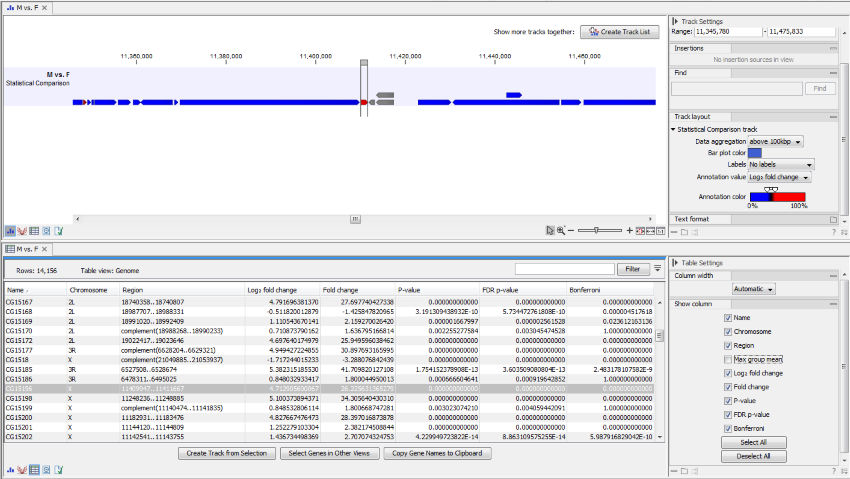
Figure 31.83: Statistical comparison track view.
In particular, the Fold Change value will tell you how expression levels in group 2 are relative to that in group 1.
- If expression values in group 2 are twice as large as in group 1, the fold change will be +2.
- If expression values in group 1 are twice as large as in group 2, the fold change will be -2.
The track layout of the statistical comparison track can be customized as follows:
- Data aggregation Allows you to specify whether the information in the track should be shown in detail or whether you wish to aggregate data. By aggregating data you decrease the detail level, but increase the speed of the data display process, which is of particular interest when working with big data sets. The threshold (in bp) for when data should be aggregated can be specified with the drop-down box. The threshold describes the unit (or "bucket") size in base pairs, above which the data will start being aggregated. The bucket size depends on the track length and the zoom level. Hence, a data aggregation threshold with a low value will only show details when zoomed in, whereas a high value means that you can see details even when zoomed out. Please note that when using the high values, it will take longer time to display the data on the screen.
- Bar plot color Selects the color of aggregated data.
- Labels Determines where the gene name should be shown.
- Annotation value The value that is graphically shown in detail view:
- Max group means For each group in the statistical comparison, the average TPM is calculated. This value is the maximum of the average TPM's.
- -log2 fold change The logarithmic fold change.
- Fold change The (signed) fold change. Genes/transcripts that are not observed in any sample have undefined fold changes and are reported as NaN (not a number).
- P-value Standard p-value. Genes/transcripts that are not observed in any sample have undefined p-values and are reported as NaN (not a number).
- FDR p-value The false discovery rate corrected p-value.
- Bonferroni The Bonferroni corrected p-value.
- Annotation color Determines how the annotation value is mapped onto a color.
The expression track table view has the following options:
- The "Filter to selection" only displays pre-selected rows in the table.
- The "Create track from Selection" will create a Track using selected rows.
- The "Select Genes in Other Views" button finds and selects the currently selected genes and transcripts in all other open expression track table views.
- The "Copy Gene Names to Clipboard" button copies the currently selected gene names to the clipboard.
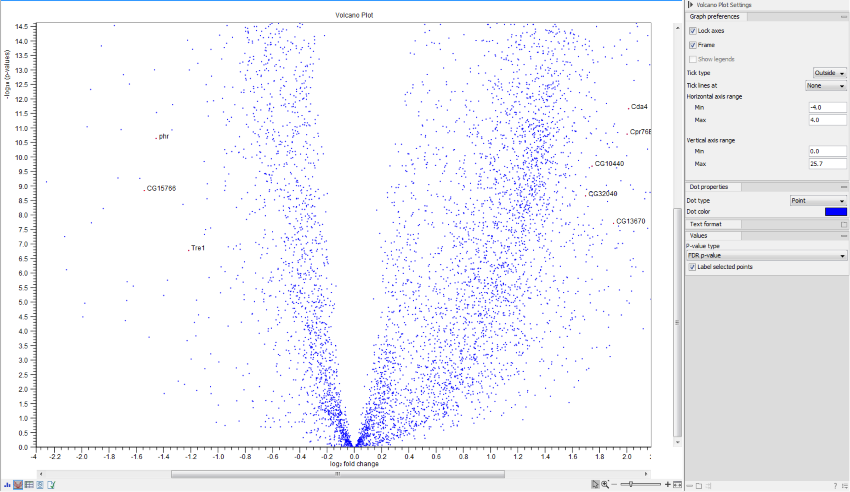
Volcano plots
Statistical comparisons also offer a volcano plot view.
An example of a volcano plot is shown in figure 31.81.
The volcano plot shows the relationship between the p-values of a statistical test and the fold changes among the samples. The log2 fold changes are plotted on the x-axis, and the -log10 p-values are plotted on the y-axis. Features of interest are typically those in the upper left and right hand corners of the volcano plot, as these have large fold changes (lie far from ![]() ) and are statistically significant (have large y-values).
) and are statistically significant (have large y-values).
Sometimes, the volcano plot will show unexpected pattern looking like "wings", such as the ones highlighted with red arrows in figure 31.82.
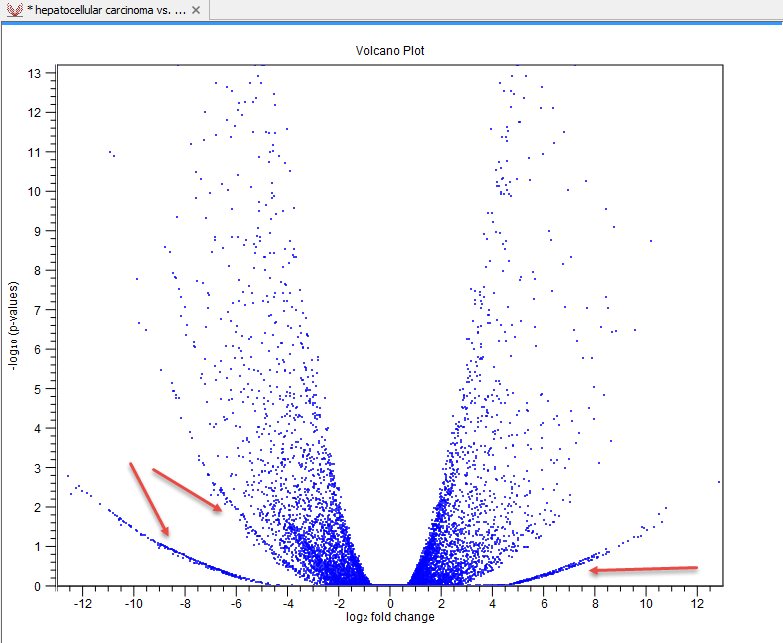
Figure 31.85: Volcano plot displaying unexpected "wing" patterns.
These patterns reflect the mathematical relationship between fold change and p-value, which often becomes exposed when there are few replicates and when expression is low in one condition. For example, expression counts for two genes might be (5,5) vs (0,0) and (5,6) vs (0,1). These two genes would appear in the same "wing". Two other genes with expression counts (5,5) vs (0,1) and (5,6) vs (0,1) might be in another "wing".
When working with several samples, it can be useful to make an Expression Browser with all the samples and to open this alongside the Volcano plot. Click a point in the Volcano plot to select it and then right-click to Select Genes in Other Views. This will select the appropriate row in the expression browser.
Volcano plot side panel It is possible to change the type of p-value from the side panel (see below).
The view settings can be adjusted using the Side Panel. Under Graph preferences, you can adjust the general properties of the volcano plot
- Lock axes This will always show the axes even though the plot is zoomed to a detailed level.
- Frame Shows a frame around the graph.
- Show legends Shows the data legends.
- Tick type Determine whether tick lines should be shown outside or inside the frame.
- Tick lines at Choosing Major ticks will show a grid behind the graph.
- Horizontal axis range Sets the range of the horizontal axis (x axis). Enter a value in Min and Max, and press Enter. This will update the view. If you wait a few seconds without pressing Enter, the view will also be updated.
- Vertical axis range Sets the range of the vertical axis (y axis). Enter a value in Min and Max, and press Enter. This will update the view. If you wait a few seconds without pressing Enter, the view will also be updated.
Below the general preferences, you find the Dot properties and Text format, where you can adjust the coloring and appearance of the dots and text.
At the bottom are options for choosing which values to display:
- P-value type Selects which type of p-value to use.
- Label selected points Chooses whether selected points should be labeled.
- Lower limit on p-values Round all p-values smaller than this number to the chosen value (for example, using the default setting, a value of zero will become 1E-16) so even small values can be visualized on a logarithmic scale volcano plot.
Note that if you wish to use the same settings next time you open a volcano plot, you need to save the settings of the Side Panel.