Illumina
CLC Genomics Workbench supports data from Illumina's Genome Analyzer, HiSeq 2000, NextSeq and the MiSeq systems. Choosing the Illumina importer opens the dialog shown in figure 6.10. This data type can also be imported using the on-the-fly import functionality described in Launching workflows individually and in batches.
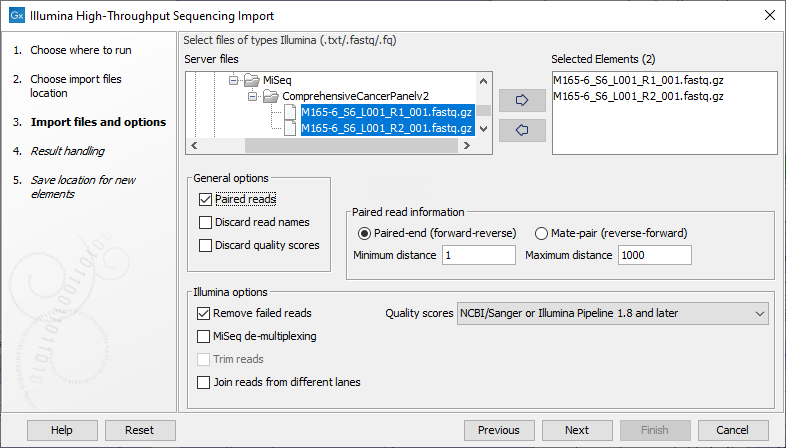
Figure 6.10: Importing data from Illumina systems.
File format
Fastq format files and fastq format files that compressed using gzip (.gz), zip (.zip) or bzip2 (.bz2) can be imported using the Illumina importer.
General Options
The settings in the General options area of the dialog are:
- Paired reads. Enable this option when importing Paired-end or Mate-pair data.
When enabled, you can specify the paired data type and the expected distance range in the "Paired read information" section of the dialog, which is described later in this section.
For paired data, pairs of files should be selected. The first reads of read pairs are expected in one file and the second reads of the pairs in another file. Multiple pairs of files can be selected. To determine which files form pairs, the files are sorted based on their names. Rules are then applied to determine whether the pairs of files are valid pairs.
Determining pairs of files
First, the selected files are sorted based on the file names. Sorting is alphanumeric, except for files coming off the CASAVA1.8 pipeline, where pairs are organized according to their identifier and chunk number.
For example, for files from CASAVA1.8, files with base names like: ID_R1_001, ID_R1_002, ID_R2_001, ID_R2_002, the files would be sorted in the order below, where it is assumed that files with names containing "R1" contain the first sequences of the pairs, and those containing "R2" in the name contain the second sequence of the pairs.
- ID_R1_001
- ID_R2_001
- ID_R1_002
- ID_R2_002
In this example, the data in files ID_R1_001 and ID_R2_001 are treated as a pair, and ID_R1_002, ID_R2_002 are treated as a pair.
The following checks are then carried out for each prospective pair of files to determine whether those files form a valid pair:
- If the file names appear to follow the following naming format:
<sample name>_L<at least one digit>_[R1|R2]_<at least one digit>, then the name of each file in the pair must have the same sample name and lane information. If they do not, no data is imported from those files and a message is printed in the log. - If the file names do not follow the naming format described above, but do contain "R1" or "R2" in their names, then the first file of the pair must contain "R1" in the name and the second file name must contain "R2". If this condition is not met, no data is imported from those files and a message is printed in the log. Note that if "R1" or "R2" appear more than once in a filename, the last instance in the name is used.
- If the file names do not match either of the cases above, then import is allowed to proceed. I.e. No further checks are done to attempt to validate if the pairs of files, as per their order in the sorted list, are a valid pair based on their filenames.
If the Join reads from different lanes option, in the Illumina options section of the dialog, is enabled, then valid pairs of files with the same lane information in their file names will be imported into the same sequence list. If a valid pair of files do not contain the same lane information in their names, then no data is imported from those files and a message is printed in the log.
Within each file, the first read of a pair will have a
1somewhere in the information line. In most cases, this will be a/1at the end of the read name. In some cases though (e.g. CASAVA1.8), there will be a1elsewhere in the information line for each sequence. Similarly, the second read of a pair will have a2somewhere in the information line - either a/2at the end of the read name, or a2elsewhere in the information line. - Discard read names. For high-throughput sequencing data, the naming of the individual reads is often irrelevant given the huge amount of reads. This option allows you to discard read names to save disk space.
Note: If you do not choose to discard read names, you can quickly check that the imported data contains the expected pairs by looking at the first few sequence names of the imported sequence list in the CLC Genomics Workbench. The first two sequences should have the same name, except for a
1or a2somewhere in the read name line. - Discard quality scores. Quality scores are visible in the mapping view and they are used during variant detection. If this is not relevant for your work, you can enable the Discard quality scores option. This can reduce disk space usage and memory consumption. Read more about the quality scores of Illumina data below.
Paired read information
When the Paired reads option is enabled, options in the "Paired read information" section of the dialog can be edited. Here, you specify the type of paired data, Paired-end (forward-reverse) or Mate-pair (reverse-forward), and the expected distance range.
In the Workbench, the only difference between paired-end (forward-reverse) or mate-pair (reverse-forward) is the expected orientation of the reads: forward-reverse in the case of paired end data and reverse-forward in the case of mate pairs.
The paired read distance includes the full read sequence, i.e. from the beginning of the forward read to the beginning of the reverse read (figure 6.11). The distances are usually defined during the library preparation of your sequencing experiment, but in doubt you can enter default values: for paired-end the distances are between 1 and 1000 bp while mate-pair reads typically have longer distances between 1000-5000 bp (and sometimes up to 10000). Note that the tools usually used subsequently to process Illumina reads (such as Map Reads to Reference or RNA-Seq Analysis) have an "Auto-detect paired distances" option that is enabled by default. As long as this option is used, mis-specifying the distances during import should bear no consequences.
Read more about handling paired data.
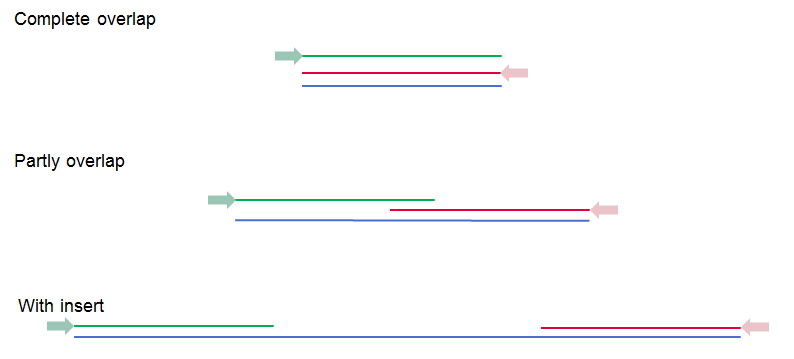
Figure 6.11: Green lines represent forward reads, red lines reverse reads, and in blue is shown the distance of the sequenced DNA fragment. Thus, if there is a complete overlap, the minimum distance will not be 0, but the length of the overlap.
Illumina options
- Remove failed reads. If you check Remove failed reads, reads that did not pass a quality filter (as indicated within the fastq files) will be ignored during import.
Part of the header information for the quality score has a flag where Y means failed and N means passed. In this example, the read has not passed the quality filter:
@EAS139:136:FC706VJ:2:2104:15343:197393 1:Y:18:ATCACG
If you import paired data and one read in a pair is removed during import, the remaining mate will be saved in a separate sequence list with single reads.
- MiSeq de-multiplexing. Using this option on MiSeq multiplexed data will divide reads into different files based on the "IndexSequence" of the read header:
@Instrument:RunID:FlowCellID:Lane:Tile:X:Y:UMI ReadNum:FilterFlag:0:IndexSequence
- Trim reads. When enabled, reads are trimmed when a B is encountered at either end of the reads in the input file. This option is only available when the "Quality score" option has been set to Illumina Pipeline 1.5 to 1.7 as a B in the quality score has a special meaning as a trim clipping in this pipeline. This trimming is carried out whether or not you choose to discard quality scores during import.
- Join reads from different lanes. When enabled, fastq files from the same sequencing run but from different lanes are imported as a single sequence list.
Lane information is expected in the filenames as "L<digits>", e.g. "L001" for lane 1. If this patterns occurs more than once in a filename, the last instance in the name is used. For example, if filenames were
myFile_L001_L1.fastqthen the lane information is taken to be L1.
In the next wizard step, options are presented for how to handle the results. If you choose to Save the results, an option called "Create subfolders per batch unit" becomes available. When that option is checked, each sequence list is saved into a separate folder under the location selected to save results to. This can be useful for organizing subsequent analysis results and for batch processing.
Subsections
