Identify mutated genes
The overlap filter will be used for filtering an annotation track based on an overlap with another annotation track. This can be used to e.g. only show variants that fall within genes or regulatory regions or for restricting variants results to only cover a subset of genes as explained in Select genes by name. Please note that for comparing variant tracks, more specific filters should be used (see Add information to variants tools).
If you are just interested in finding out whether one particular position overlaps any of the annotations, you can use the advanced table filter and filter on the region column (track tables are described in Showing a track in a table).
Toolbox | Identify Candidate Genes (![]() ) | Identify Mutated Genes (
) | Identify Mutated Genes (![]() )
)
Select the track you wish to filter and click Next to specify the track of overlapping annotations (see figure 26.5).
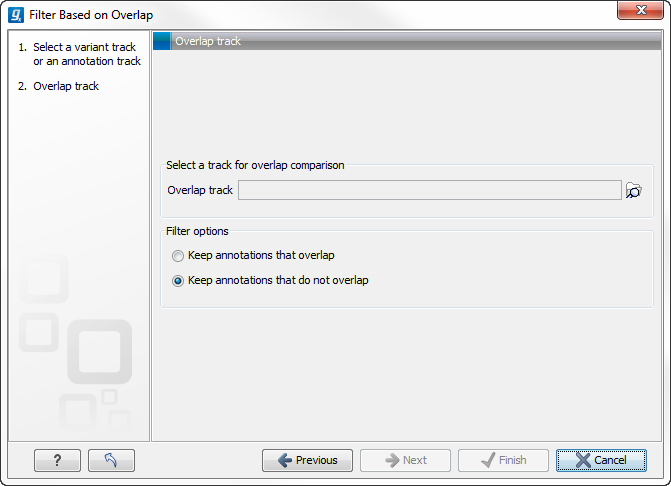
Figure 26.5: Select overlapping annotations track.
Next, select the track that should be used for comparison and tick whether you wish to keep annotations that overlap, or whether to keep annotations that do not overlap with the track selected. An overlap has a simple definition - if the annotation used as input has at least one shared position with the other track, there is an overlap. The boundaries of the annotations do not need to match.
