Quality-based variant detection
The quality-based variant detection in CLC Cancer Research Workbench is based on the Neighborhood Quality Standard (NQS) algorithm of [Altshuler et al., 2000] (also see [Brockman et al., 2008] for more information).
Using a combination of quality filters and user-specified thresholds for coverage and frequency, this tool finds all variants that are covered by aligned reads.
To run the variant detection:
Toolbox | Legacy Tools (![]() ) | Quality-based Variant Detection (legacy) (
) | Quality-based Variant Detection (legacy) (![]() )
)
This opens a dialog where you can select mapping results (![]() )/ (
)/ (![]() )/ (
)/ (![]() ) or RNA-Seq analysis results (
) or RNA-Seq analysis results (![]() ).
).
Clicking Next will display the dialog shown in figure 32.1
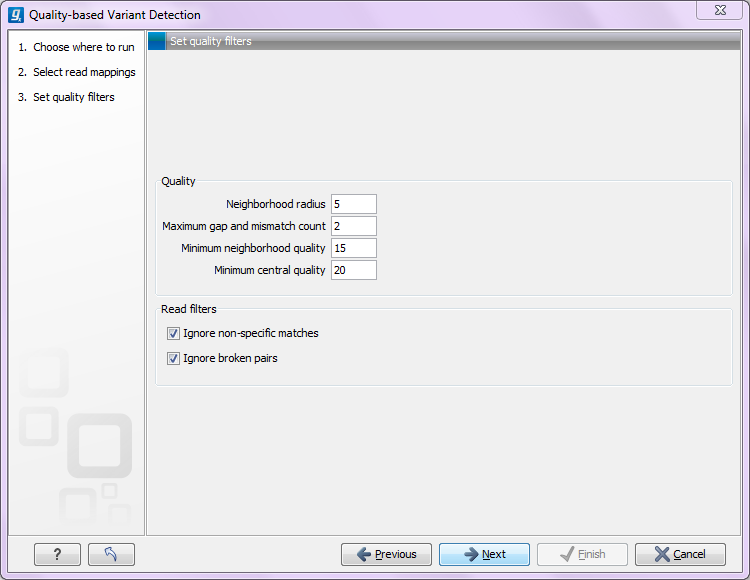
Figure 32.1: Quality filtering.
Subsections
- Assessing the quality of the neighborhood bases
- Significance of variant
- Ploidy and genetic code
- Reporting the variants
