The Perform QIAseq cfDNA Analysis template workflow
The Perform QIAseq cfDNA analysis workflow supports analysis of Illumina QIAseq cfDNA panel data. The cfDNA panels are designed to provide high coverage in targeted regions to allow identification of low frequency variants. As the read structure is different from standard QIAseq panels, the Perform QIAseq cfDNA analysis workflow should only be used to process data from from QIAseq cfDNA panels.
The Perform QIAseq cfDNA analysis workflow is set up to detect very low frequency variants. Please note that to call low frequency variants, coverage must be high. In low coverage samples or regions, very low frequency variants are unlikely to be represented in the reads.
The primers and ROIs for cfDNA panels are not yet available in the CLC reference data, but the workflow is pre-configured with all other input files such hg19 reference sequence, mRNA etc.
To run the workflow go to:
Template Workflows | Biomedical Workflows (![]() ) | QIAseq Panel Analysis (
) | QIAseq Panel Analysis (![]() ) | QIAseq Analysis Workflows (
) | QIAseq Analysis Workflows (![]() ) | The Perform QIAseq cfDNA analysis (
) | The Perform QIAseq cfDNA analysis (![]() )
)
You can then select the reads to analyze (figure 6.42).
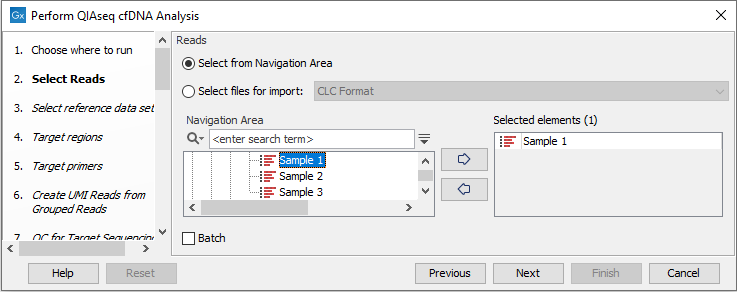
Figure 6.42: Choose the reads sequenced with a cfDNA panel.
In the next dialog, check "Use the default reference data" to allow specification of relevant target regions and primers in the following wizard steps (figure 6.43).
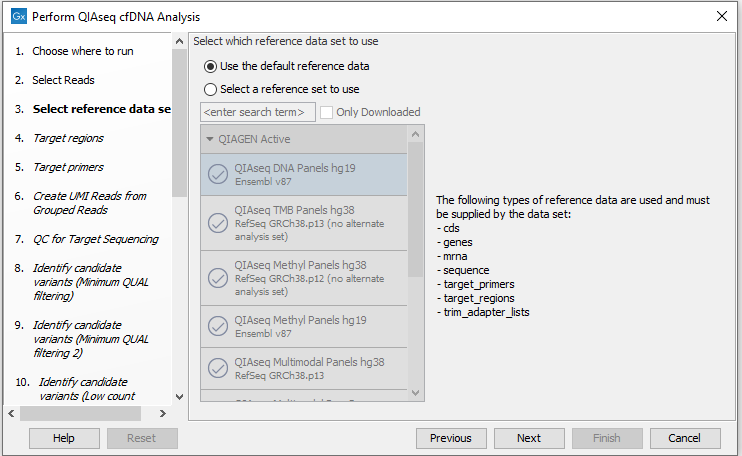
Figure 6.43: Choose the appropriate Reference Data Set.
In the Target regions dialog, select the panel specific target regions track (figure 6.44). Variants will only be detected within target regions.
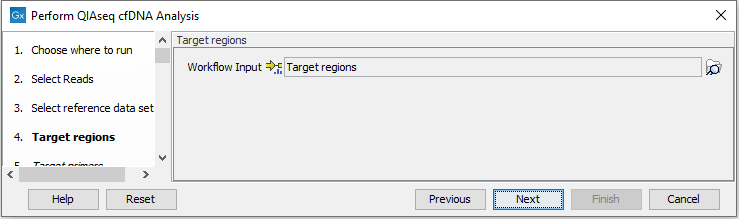
Figure 6.44: Choose the relevant Target regions track.
Repeat the selection of the appropriate track for Target primers in the subsequent dialog.
In the Create UMI Reads from Grouped Reads dialog, it is possible to specify settings for UMI gropuing (figure 6.45).
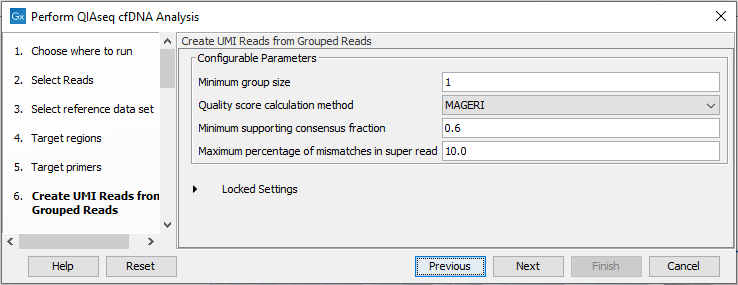
Figure 6.45: Configuring the settings for creation of UMI reads.
If the cfDNA data contains very large UMI groups, more PCR or sequencing errors than expected in normal sequencing protocols may be present in the UMIs. Therefore, settings for grouping reads into UMI reads should be more relaxed than settings for standard panels. This is reflected in the default settings for Create UMI Reads from Groups Reads in this workflow. See Create UMI Reads from Grouped Reads for details about UMI grouping using the tool Create UMI Reads from Grouped Reads.
In the QC for Target Sequencing dialog, specify the minimum coverage for QC (figure 6.46). Using default settings, samples where 90 percent of target region positions do not meet this threshold will be flagged in the Combined report generated by the workflow.
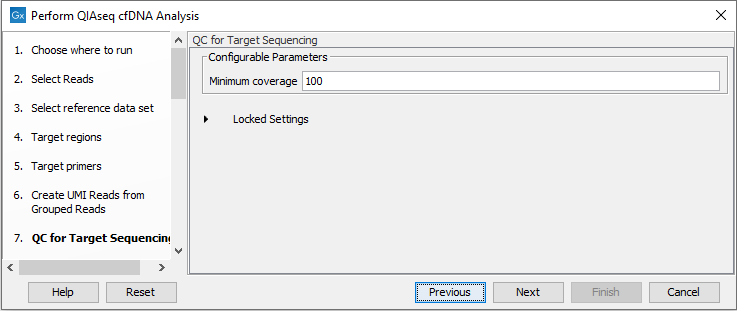
Figure 6.46: Configuring the QC for Target Sequencing tool.
Next, set the variant quality filtering thresholds QUAL and Average quality (figure 6.47). Variants with quality below the specified thresholds will not be reported.
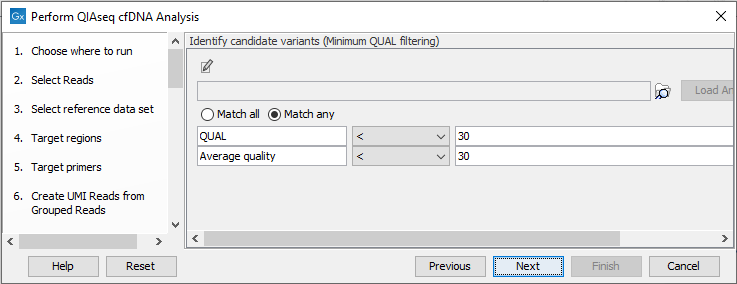
Figure 6.47: Specify quality threshold for filtering variants.
In the following wizard steps, it is possible to specify additional variant filtering criteria. When each of these have been set, choose where to save the results and press Finish to start the analysis.
Subsections
