Annotating a reference genome with genes and transcripts
In this example we use the horse genome but the methods described here apply equally well for other genomes. First, we download the fasta files for the reference genome at Ensembl: ftp://ftp.ensembl.org/pub/current_fasta/equus_caballus/dna/. The whole genome can be downloaded as a single file that ends with.dna.toplevel.fa.gz. Import (To annotate the reference with the genes and transcripts from the GTF file:
From the CLC Main Workbench:
Toolbox | General Sequence Analysis (![]() )|
Annotate with GFF/GTF File (
)|
Annotate with GFF/GTF File (![]() )
)
From the CLC Genomics Workbench:
Toolbox| Classical Sequence Analysis (![]() )| General Sequence Analysis (
)| General Sequence Analysis (![]() )| Annotate with GFF/GTF File (
)| Annotate with GFF/GTF File (![]() )
)
Now, select the horse chromosomes and click Next. This opens the dialog shown in figure 2.1.
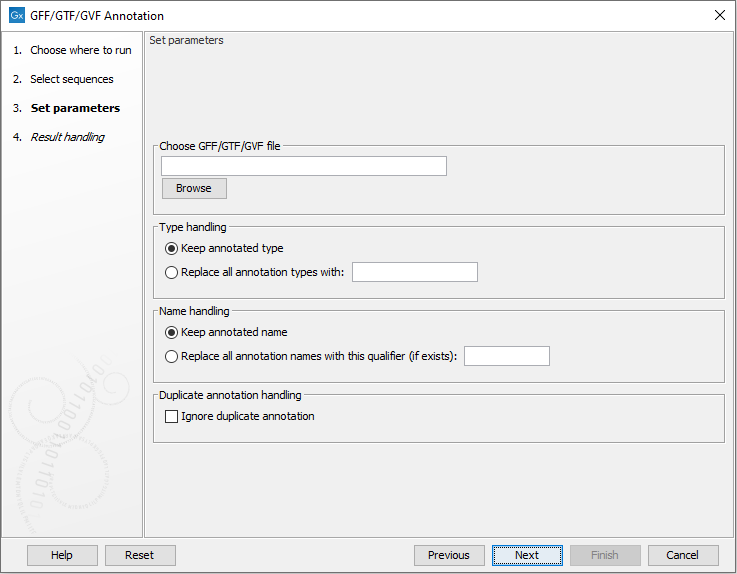
Figure 2.1: Select the GTF file by clicking the browse icon.
Click Browse to select the GFF/GTF file and click Next. Choose to Save the results and click Finish. This will add the annotations from the file to the sequences. Your reference genome is now ready for use.
Notes about gene annotations from the UCSC. GTF-files downloaded from the UCSC genome browser are not compatible with choosing to run RNA-Seq Analysis on a annotated eukaryotic reference because the gene and transcript annotations cannot be matched. You may choose to use USCS gene annotations only for RNA-Seq analysis: In the CLC Genomics Workbench version 7.x you can choose to only consider gene annotations by choosing the option "Genome annotated with genes only". For the CLC Genomics Workbench version 6.5.x and earlier, you can get the same effect by choosing to treat the reference as an annotated prokaryotic reference.
We would, however, generally recommend getting the annotations from a source where genes and transcripts are linked for the purposes of RNA-Seq on eukaryotic genomes, such as from Ensembl.
