Data QC and OTU Clustering workflow
The Data QC and OTU Clustering workflow consists of 5 tools being executed sequentially (figure 4.10). The only necessary input to run the workflow are the reads you want to cluster. You also have the option to provide a list of the primers that were used to sequence these reads if you wish to perform the adapters trimming step with the Trim Sequences tool.
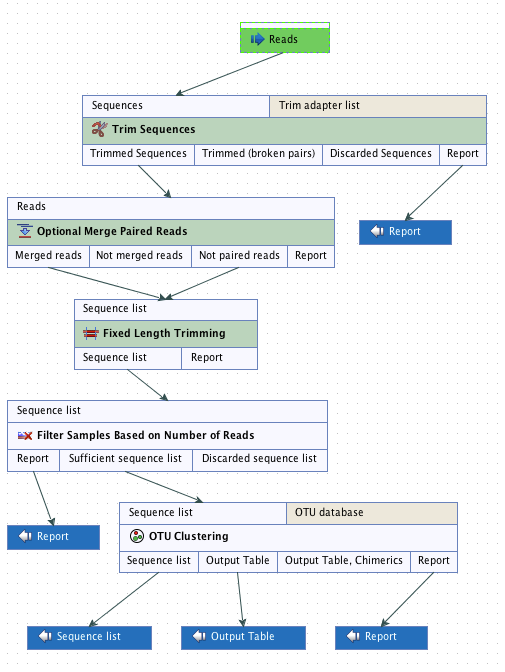
Figure 4.10: Layout of the Data QC and OTU clustering workflow.
The first tool is the Optional Merge Paired Reads that will output 2 sets of sequences, the merged reads and the not paired reads. Both will be used as input in the Trim Sequences tool together with the sequencing primer list. This tool provides a list of trimmed sequences that will be the input of the Fixed Length Trimming tool. Again, the output is a list of trimmed sequences, used as input file for the Filter Samples Based on the Number of Reads tool. The results of the filter are compiled in a report and the tool generates a sequence list that does not contain the reads of poor quality. This "filtered" list will be used for the final tool of the workflow, the OTU clustering tool. This tool will give 2 outputs: a sequence list of the OTU centroids and an abundance table with the newly created OTUs, their abundance at each site as well as the total abundance for all samples.
