Running Pfam Domain Search
When you have downloaded the Pfam database you are ready to perform a Pfam domain search. To do this start the Pfam search tool:
Toolbox | Classical Sequence Analysis (![]() ) | Protein Analysis (
) | Protein Analysis (![]() )| Pfam Domain Search (
)| Pfam Domain Search (![]() )
)
If a sequence was selected before choosing the Toolbox action, this sequence is now listed in the Selected Elements window of the dialog. Use the arrows to add or remove sequences or sequence lists from the selected elements.
You can perform the analysis on several protein sequences at a time. This will add annotations to all the sequences. Click Next to adjust parameters (see figure 15.16).
Figure 15.16: Setting parameters for Pfam Domain Search.
- Database. Choose which database to use when searching for Pfam domains. For information on how to download a Pfam database see .
- Significance cutoff
- Use profile's gathering cutoffs. Use cutoffs specifically assigned to each family by the curator instead of manually assigning the Significance cutoff.
- Significance cutoff. The E-value (expectation value) describes the number of hits one would expect to see by chance when searching a database of a particular size. Essentially, a hit with a low E-value is more significant compared to a hit with a high E-value. By lowering the significance threshold the domain search will become more specific and less sensitive, i.e. fewer hits will be reported but the reported hits will be more significant on average.
- Remove overlapping matches from the same clan. Perform post-processing of the results where overlaps between hits are resolved by keeping the hit with the smallest e-value.
Click Next to adjust the output of the tool. The Pfam search tool can produce two types of output. It can add annotations on the input sequences that show the domains found (see figure 15.17) and it can output a table with all the domains found.
Click Next if you wish to adjust how to handle the results. If not, click Finish.
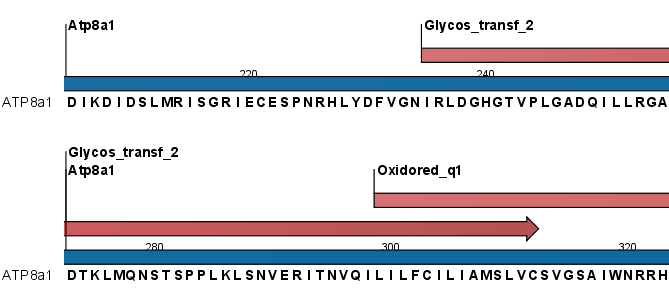
Figure 15.17: Annotations (in red) that were added by the Pfam search tool.
Domain annotations added by the Pfam search tool have the type Region. If the annotations are not visible they have to be enabled in the side panel. Detailed information for each domain annotation, such as the bit score which is the basis for the prediction of domains, is available through the annotation tool tip.
A more detailed description of the scores provided in the annotation tool tips can be found here: http://pfam.sanger.ac.uk/help#tabview=tab5.
