Sample reads
The Sample Reads tool allows a user defined reduction of the number of reads. This can be relevant when editing contigs with a high coverage as an excessive amount of reads can be overly resource demanding and cause the edit options in the read mapping editor to disallow performing actions. The Sample Reads tool is very useful for reducing the size of data sets. The sampled reads can sequentially be mapped to contigs in the Contig Aligner. In this way the speed of many edit operations in the contig aligner can be increased significantly without any loss of important information. The preservation of a high coverage is only necessary for de novo assembly. After de novo assembly a large number of reads does not add extra to the analysis.
How to run Sample Reads
Toolbox | NGS Core Tools (![]() ) | Sample reads (
) | Sample reads (![]() )
)
This opens the dialog shown in figure 24.54.
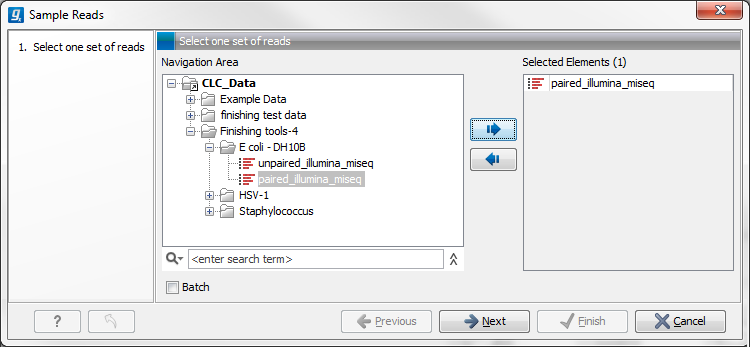
Figure 24.54: Select the reads.
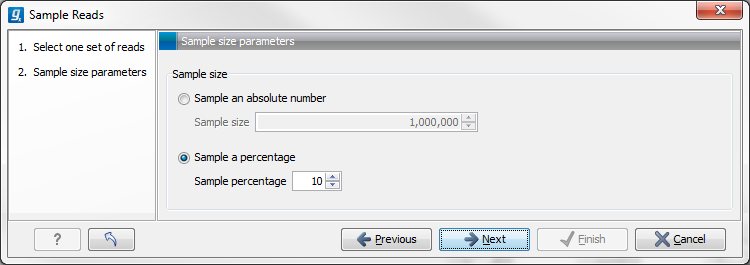
Figure 24.55: Specify the sample size parameters.
Select the relevant assembled reads and click Next. This leads to the Sample size parameters step shown in figure 24.55.
In the Sample size parameters it is possible to select either an absolute number of reads to sample or a sample percentage, with sample percentage defining the percentage of the sub-sample paired reads (for single end reads) and pairs of reads (for paired reads).
After the Result handling step, click Finish.
This generates a sampled version of the reads that can be used in a new read mapping or used to replace existing reads in the Contig Aligner.
