Probabilistic variant detection
The purpose of the Probabilistic Variant Caller is to identify variants in a sample by using a probabilistic model built from read mapping data. This tool can detect variants in data sets from haploid (e.g. Bacteria), diploid (e.g. Human) and polyploid organisms (e.g. Cancer and higher plants) with a high sensitivity and specificity.
The algorithm used is a combination of a Bayesian model and a Maximum Likelihood approach to calculate prior and error probabilities for the Bayesian model.
Parameters are calculated on the mapped reads alone without considering the reference sequence. After observing a certain combination of nucleotides from the reads at every position in the genome, the probability for each combination of alleles (e.g. homozygous A/A, heterozygous A/G, heterozygous A/C etc.) will be determined. This probability is then used to find out which of the allele combinations (e.g. A/G) is the most likely one for each position. This can then be compared with the reference allele to find out if it is different from the reference sequence and therefore can be called as a variant.
Note: In the current version, the probabilistic variant detection is not designed to detect minor variants (like rare alleles) with a frequency of less than 15%. If you are expecting a allele frequency of less than 15% we would recommend setting a higher ploidy level during your analysis or alternatively, using the quality-based variant detection algorithm (see Quality-based variant detection) with a post-filtering step for average base quality and forward-reverse read balance.
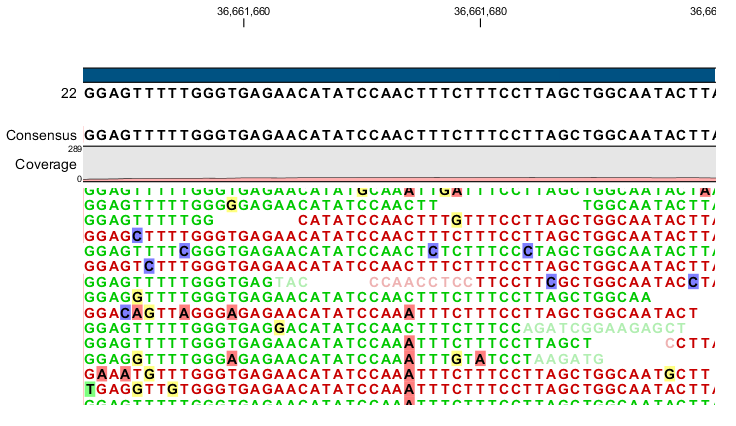
Figure 26.12: An example of a heterozygous variant surrounded by a lot of noise from sequencing errors.
Subsections
- Calculation of the prior and error probabilities
- Calculation of the likelihood
- Calculation of the posterior probability for each site type at each position in the genome
- Comparison with the reference sequence and identification of candidate variants
- Posterior filtering and reporting of variants
- Running the variant detection
- Setting ploidy and genetic code
- Reporting the variants found
