Import Expression Data
Import Expression Data enables import of individual expression tracks from an expression data matrix. The data matrix needs to be constructed to conform with the following formatting:
- The matrix should be constructed in Excel or csv format.
- Columns represent samples and rows represent genes. See figure 3.1 for an example of correct formatting.
- Feature ID (gene ID or transcript ID) should be in the first column and samples in the following.
- Only one feature ID is supported. It should be unique, i.e. Ensembl or geneID, not a mixture. Features that are non-unique will be ignored.
- Three types of expression values are supported: Raw counts, TPM, an RPKM. Only one of these values should be supplied. We recommend to use raw counts when available.
- If the matrix has been filtered for low count entries before upload, the provided calculation of TPM or RPKM needs to be on the filtered matrix as well, otherwise the counts will not be properly translated.
- Import of other normalization types are not supported.
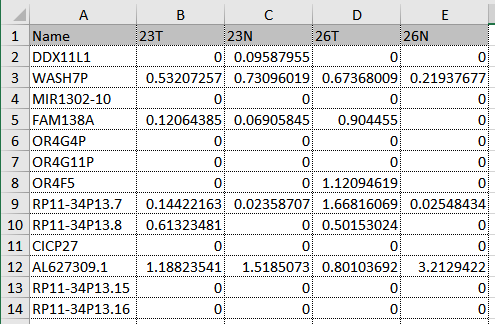
Figure 3.1: RPKM count matrix using Ensembl gene names and representing 4 samples in a Tumor Normal design.
To launch the Import Expression Data tool, go to:
Toolbox | Ingenuity Pathway Analysis | Import Expression Data
Figure 3.2 shows the Import Expression Data dialog.
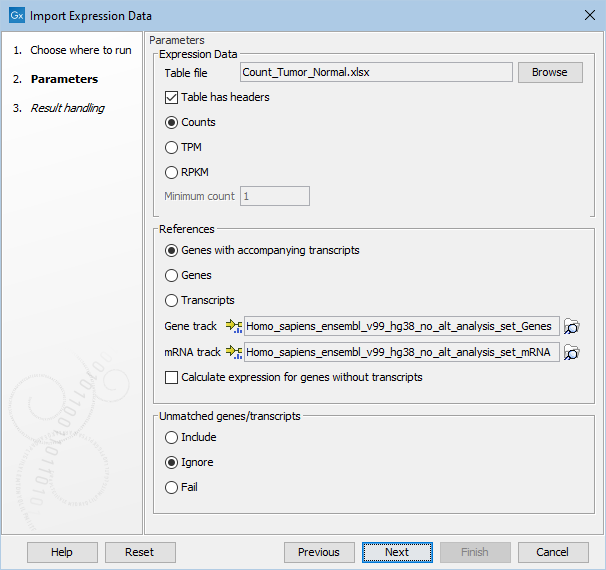
Figure 3.2: Parameters available in the Import Expression Data tool. Select the Table file containing the expression matrix and select the type of data matching the values in the file (in this case it contains count data). Add references to import against appropriate gene or transcript annotations. Select how to handle unmatched genes or transcripts.
In the Expression Data section of the dialog that opens, first select the data matrix by using the Browse button.
Select the expression values that matches the expression data type. All value types must be non-negative values:
- Counts
- TPM
- RPKM
When selecting TPM or RPKM, the expected minimum count must be specified. The value must be the smallest count value that was present in the expression matrix when calculating the TPMs or RPKMs values. In unfiltered data this value will typically be 1 (default).
Under References, specify how expression values were generated. This is for defining whether it was generated as a gene or transcript matrix as well as to specify how the TPM/RPKM were calculated.
- Genes with accompanying transcripts Matches imported values against genes. Transcripts are used for identifying exon length when translating between counts and TPM/RPKM.
- Genes Matches imported values against genes. Gene length are used when translating between counts and TPM/RPKM.
- Transcripts Matches imported values against transcripts and uses exon length when translating between counts and TPM/RPKM.
The key is that you specify the Gene and mRNA tracks that were used to generate the expression values. When selecting Genes with accompanying transcripts as parameter you can choose to calculate expression for genes without transcript. This will result in the generation of a transcript that is expected to have the length of the full gene. Enabling this option allows calculation of TPM and RPKM when counts have been supplied.
At the bottom of the dialog, specify whether unmatched genes or transcripts should be included or ignored, or if the analysis should fail.
The Import Expression Data tool outputs one expression track per samples.
Subsections
