Update Sequence Attributes in Lists
Update Sequence Attributes in Lists updates information pertinent to sequences within a sequence list. For example, descriptions can be updated, or new information types can be added. The attribute information to add to the sequences within a sequence list is provided via an Excel file. The attribute/column to use to match upon, so that information is the relevant row is added to a particular sequence, is specified when launching the tool.
Attribute information for each sequence can be seen when viewing a sequence list in table view. Many attributes can be directly edited, or updated using the Update Sequence Attributes in Lists tool. Some, however, cannot be, for example the length or the start of the sequence, as these are characteristics of the sequence itself.
Individual values can be updated manually by right-clicking in the relevant cell and choosing to edit that attribute (figure 25.3). Working with editable attributes in tables is described in Working with tables.
Alternatively, right click on an individual sequence in the sequence list and choose to open that sequence. Then navigate to the Element info view and change attribute values there. Changes made in the Element info view are reflected immediately in the open sequence list.
For updating information for many sequences, the Update Sequence Attributes in Lists is recommended.
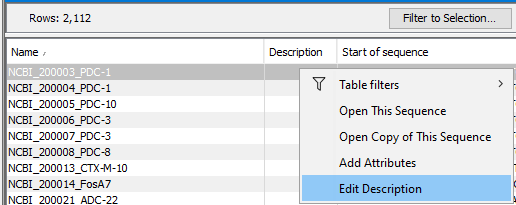
Figure 25.3: Attributes on individual sequences in a sequence list can be updated. Right click in the relevant cell in table view, and choose to edit that attribute.
To launch the Update Sequence Attributes in Lists tool, go to:
Toolbox | Utility Tools (![]() ) | Sequence Lists (
) | Sequence Lists (![]() ) |Update Sequence Attributes in Lists (
) |Update Sequence Attributes in Lists (![]() )
)
and select a sequence list as input (figure 25.4).
Multiple sequence lists of the same type (nucleotide or peptide) can be selected as input, however please note that sequences in all lists are considered together as a single input, and a single sequence list will be output, containing all sequences of the input lists.
Figure 25.4: Select a sequence list as input to the tool.
In the second wizard step, the source of the attribute information is specified, along with details about how to handle that information.
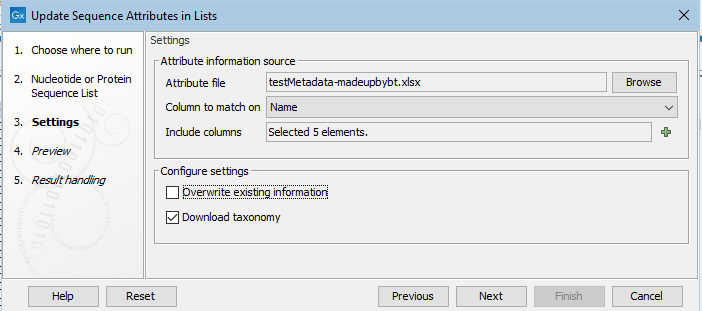
Figure 25.5: Attributes from 5 columns in the specified file will be added or updated. Existing information will not be overwritten. If one of the specified columsn is called TaxID, then a 7-step taxonomy will be downloaded from the NCBI and added to an attribute called Taxonomy.
Attribute information source
- Attribute file An Excel (.xlsx) file containing attribute information, with a header row containing the names of attributes.
- Column to match on The specified column heading will be matched against a sequence attribute name. When a row in that column is identical to the value for that attribute in one or more sequences, the information from the Excel sheet is added to those sequences. If there are columns present for attribute types not already defined for the sequence list, that attribute type is added.
- Include columns The columns from the Excel file containing attributes to be added to the sequence list. If the "Download taxonomy" option, described below, is checked, a column called Taxonomy will be assumed to included, and will be listed in the preview shown in the next step.
Configure settings
- Overwrite existing information When checked, if there is a new value for an existing attribute, the old value will be overwritten by the new value. When unchecked existing values remain, without change, whether or not a new value is present in the Excel file.
- Download taxonomy When checked, a column called TaxIDs is expected, containing valid taxonomic identifiers. A 7-step taxonomy is then downloaded from the NCBI into an attribute called "Taxonomy".
Examples of valid identifiers for TaxID attribute are those found in
/db_xref="taxonfields in Genbank entries. For example, for/db_xref="taxon:5833, the expected value in the TaxID column would be5833.If a given sequence has an value already set for the Taxonomy attribute, then that existing value remains in place unless the "Overwrite existing information" box is checked.
The next step provides a preview of the updates that will be made. In the upper pane, a list of the attribute types to be considered is listed. For certain attribute types, recognized by particular column names, validation rules are applied. For example, a column named GO-terms is expected to contain terms in the format, GO:<id>, e.g. GO:0046782. For these, the attribute values, as seen in table view, will be hyperlinked to the relevant GO entry online at http://amigo.geneontology.org.
This list of column headings recognized in this way, and how the values in those columns is handled, is described below.
In the bottom pane, attribute values that will be added are shown for a small subset of sequences. If these are not as expected, clicking on the "Previous" button takes you back to the previous step, where the configuration can be updated.
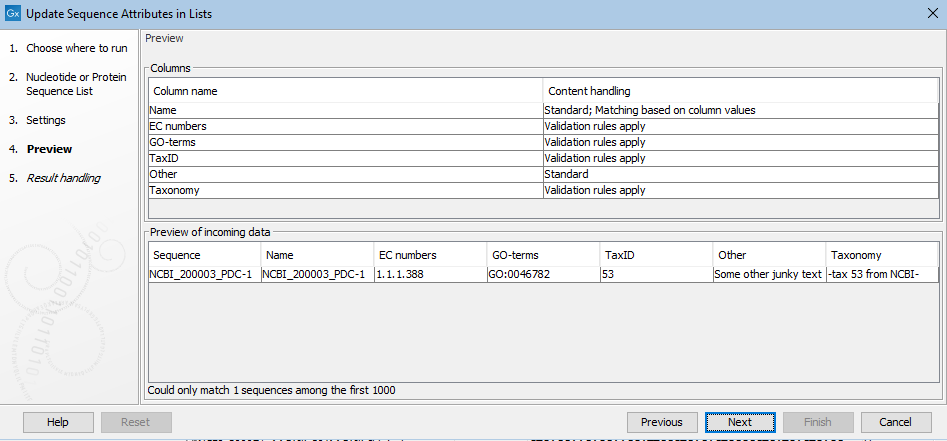
Figure 25.6: Attributes from several columns are subject to validation checks. If any had failed the check, a yellow exclamation mark in the bottom pane would be shown for that column. Here, all entries pass. The "Other" column is not subject to validation checks. Only one sequence in the list is being updated in this example.
Column headings and value validation
Certain column headings are recognized, and if the contents pass validation rules, the entries are handled by the software, generally adding hyperlinks to an online data resources.
Two columns subject to validation have additional handling:
- TaxID When valid taxonomic identifiers are found in a TaxID column, and the Download taxonomy checkbox is enabled, then a 7-step taxonomy is then downloaded from the NCBI. This is described further up on this page.
- Gene ID The following identifiers in a Gene ID column are added as attribute values and hyperlinked to the relevant online database:
- Ensembl Gene IDs
- HUGO Gene IDs
- VFDB Gene IDs
Multiple identifiers in a given cell, separated by commas, will be added as multiple Gene ID attributes for the relevant sequence. If any one of those identifiers is not recognized as one of the above types, then none will be hyperlinked.
Other columns where contents are validated are those with the headings listed below. If a value in such a column cannot be validated, it is not added nor used to update attributes.
If you wish to add information of this type but do not want this level of validation applied, use a heading other than the ones listed below.
- COG-terms COG identifiers
- Compound AROs Antibiotic Resistance Ontology identifiers
- Compound Class AROs Antibiotic Resistance Ontology identifiers
- Confers-resistance-to ARO Antibiotic Resistance Ontology identifiers
- Drug ARO Antibiotic Resistance Ontology identifiers
- Drug Class ARO Antibiotic Resistance Ontology identifiers
- EC numbers EC identifiers
- GenBank accession Genbank accession numbers
- Gene ARO Antibiotic Resistance Ontology identifiers
- GO-terms Gene Ontology (GO) identifiers
- KO-terms KEGG Orthology (KO) identifiers
- Pfam domains PFAM domain identifiers
- Phenotype ARO Antibiotic Resistance Ontology identifiers
- PubMed IDs Pubmed identifiers
- TIGRFAM-terms TIGRFAM identifiers
- Virulence factor ID Virulence Factors of Pathogenic Bacteria identifiers
Updating Location-specific attributes
Location-specific attributes can be created, which are the present for all elements created in that CLC File Location. Such attributes can be updated using the Update Sequence Attributes in Lists tool.
Of note when working with such attributes:
- Because these attributes are tied to the Location, they will not appear until the updated sequence list has been saved.
- The updated sequence list must be saved to the same File Location as the input for these attributes and their values to appear.
- If this tool is run on an unsaved sequence list, or using inputs from more than one File Location at the same time, Location-specific attributes will not be updated. Information in the preview pane reflects this.
Location-specific attributes are described in Customized attributes on data locations.
