Tracks (see Tracks) are imported in a special way, because extra information is needed in order to interpret the files correctly.
Tracks are imported using:
click Import (![]() ) in the Toolbar | Tracks
) in the Toolbar | Tracks
This will open a dialog as shown in figure 6.15.
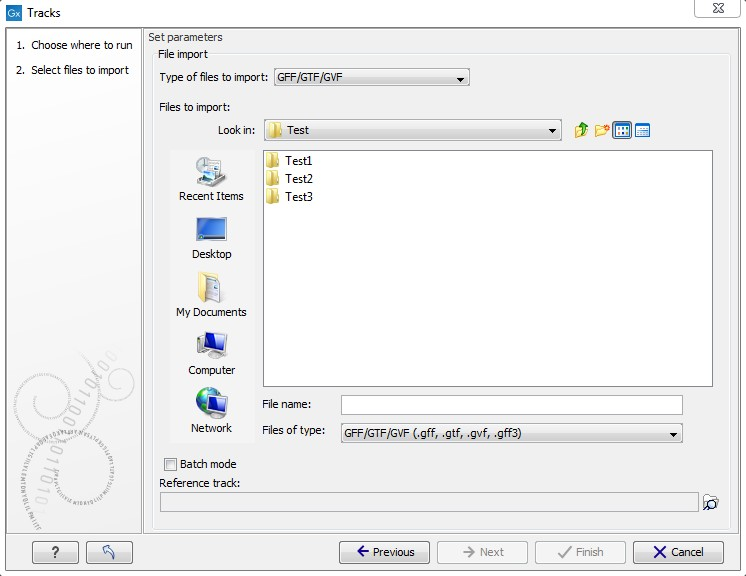
Figure 6.15: Define the reference genome.
At the top, you select the file type to import. Below, select the files to import. The formats currently accepted are:
http://www.clcbio.com/clc-plugin/annotate-sequence-with-gff-file/. This can be particularly useful when working with transcript annotations downloaded from from Ensembl available in gvf format: http://www.ensembl.org/info/data/ftp/index.html.
.txt.gz on this list can be used:
http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/. Please
note that importer is for variant data and is not a general importer for all
annotation types. This is mainly intended to allow you to import the popular
Common SNPs variant set from UCSC. The file can be downloaded from the
UCSC web site here:
http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/snp132Common.txt.gz.
Other sets of variant annotation can also be downloaded in this format using
the UCSC Table Browser.
If you want to import several files into a single track in one step you can use the batch mode function (see figure 6.15). Please be aware that this is not possible if you work with VCF files without genotype information.
Please see Annotation and variant formats for more information on how different formats (e.g. VCF and GVF) are interpreted during import in CLC format.
For all of the above, zip files are also supported.
Please note that for human data, there is a difference between the UCSC genome build and Ensembl/NCBI for the mitochondrial genome. This means that for the mitochondrial genome, data from UCSC should not be mixed with data from other sources (see http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg19).
Most of the data above is annotation data and if the file includes information about allele variants (like VCF, Complete Genomics and GVF), it will be combined into one variant track that can be used for finding known variants in your experimental data. When the data cannot be recognized as variant data, one track is created for each annotation type.
For all types of files except fasta, you need to select a reference track as well. This is because most the annotation files do not contain enough information about chromosome names and lengths which are necessary to create the appropriate data structures.