The Quantify QIAseq RNA Expression template workflow
The Quantify QIAseq RNA Expression template workflow can be found here:
Template Workflows | Biomedical Workflows (![]() ) | QIAseq Sample Analysis (
) | QIAseq Sample Analysis (![]() ) | Quantify QIAseq RNA Expression (
) | Quantify QIAseq RNA Expression (![]() )
)
Before running the workflow, make sure proper adapter trimming is performed. Run QC for Sequencing Reads to assess the reads and remove any unexpected adapters before running the workflow. Reads with adapters are likely not to map to the reference which can lead to loss of read count.
Double-click on the Quantify QIAseq RNA Expression template workflow to run the analysis.
If you are connected to a CLC Server via your Workbench, you will be asked where you would like to run the analysis. We recommend that you run the analysis on a CLC Server when possible.
The workflow can and should be run in batch mode, allowing the analysis of several samples at once. Once you have checked the Batch option, you can select the folder holding the samples that should be analyzed (figure 10.1).
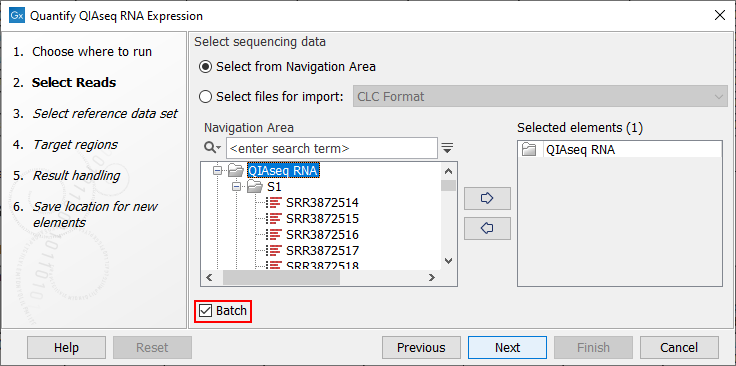
Figure 10.1: Select the samples to analyze by working in batch mode and choosing the top folder holding all samples.
When working in batch mode, it is important to select the folder containing the samples, and not the subfolders containing the sequence lists, nor the reads themselves. In the two latter cases, each sequence list would be considered as an independent sample, when in fact, individual samples are usually made of several sequence lists.
The following dialog helps you set up the relevant Reference Data Set. If you have not downloaded the Reference Data Set yet, the dialog will suggest the relevant data set and offer the opportunity to download it using the Download to Workbench button. Select QIAseq RNA Panels hg38 and Download the set if you have not done so before (figure 10.2).
Note that if you wish to Cancel or Resume the Download, you can close the template workflow and open the Reference Data Manager where the Cancel, Pause and Resume buttons are available.
If the Reference Data Set was previously downloaded, the option "Use the default reference data" is available and will ensure the relevant data set is used. You can always check the "Select a reference set to use" option to be able to specify another Reference Data Set than the one suggested.
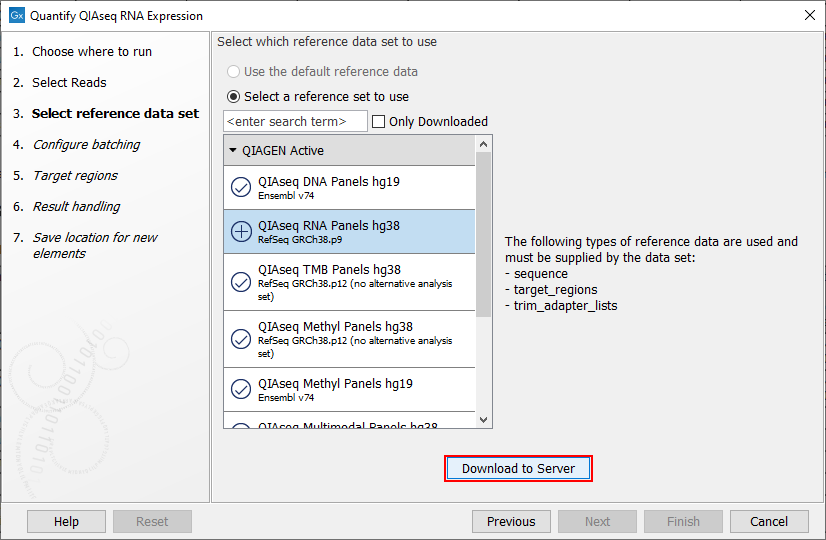
Figure 10.2: The relevant Reference Data Set is highlighted. In the text to the right, the types of reference needed by the workflow are listed.
In the next dialog you can can choose to use metadata to configure the batching if you do not want to use the default organization of input.
The batch overview dialog that comes next in the wizard allows you to check that the batch unit is the sample, as opposed to independent sequence lists. You can take advantage of this dialog to exclude some samples from your analysis (figure 10.3).
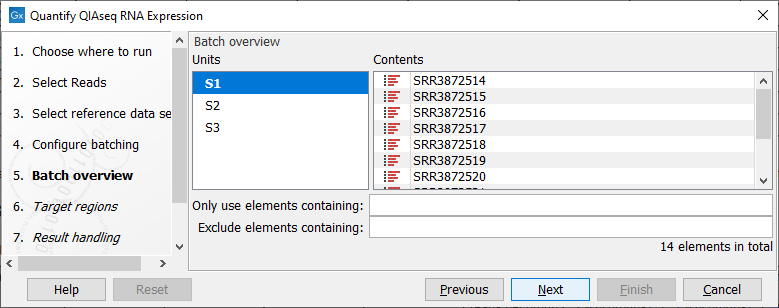
Figure 10.3: Check in this dialog that the batch unit is the sample and not the reads.
In the Target regions dialog, specify the Target regions file that correspond to the panel used from the drop down list (figure 10.4).
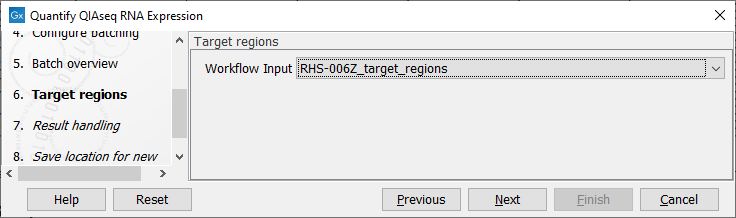
Figure 10.4: Select the Target regions track that correspond to the panel used.
Finally, in the last wizard step, choose to Save the results of the workflow before clicking Finish.
Subsections
