Identify Variants (WGS)
The Identify Variants (WGS) tool takes sequencing reads as input and returns identified variants in a Track List.
The tool runs an internal workflow that first maps the sequencing reads to the human reference sequence. Next, it runs a local realignment that is used to improve the variant detection that comes after the local realignment.
Two different variant callers are used; the Low Frequency Variant Detection tool is used to call small insertions, deletions, SNVs, MNV, and replacements, and the InDel and Structural Variants tool calls larger insertions, deletions, translocations, and replacements. By the end of the variant detection, variants that have been detected by the Low Frequency Variant Detection tool with an average base quality smaller than 20 are filtered away.
A detailed mapping report is created to inspect the overall coverage and mapping specificity in the targeted regions.
Run the Identify Variants (WGS) workflow
To run the Identify Variants (WGS) workflow, go to:
Ready-to-Use Workflows | Whole Genome Sequencing (![]() ) | Somatic Cancer (
) | Somatic Cancer (![]() ) | Identify Variants (WGS) (
) | Identify Variants (WGS) (![]() )
)
- Select the sequencing reads from the sample that should be analyzed (figure 20.24).
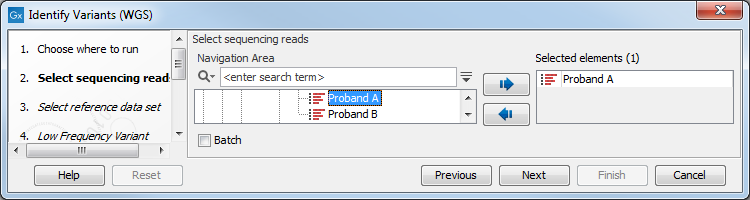
Figure 20.24: Please select all sequencing reads from the sample to be analyzed.If several samples should be analyzed, the tool has to be run in batch mode. This is done by checking "Batch" and selecting the folder that holds the data you wish to analyze.
- In the next dialog, you have to select which data set should be used to identify variants (figure 20.25).
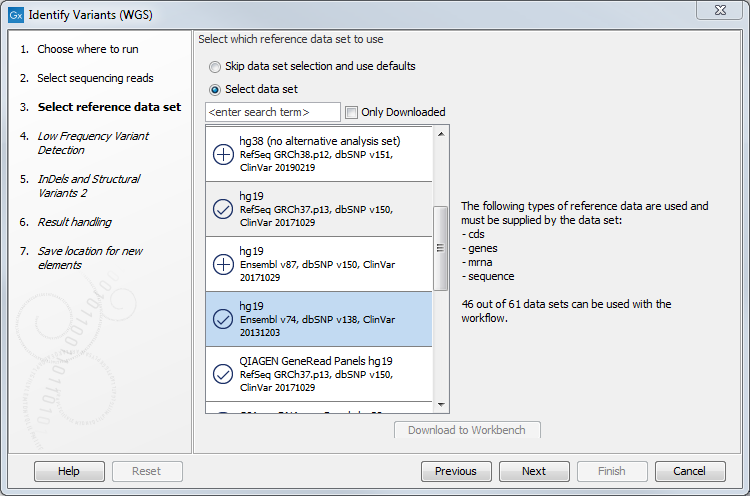
Figure 20.25: Choose the relevant reference Data Set to identify variants in your sample. - In the Low Frequency Variant Detection dialog (figure 20.26), you can specify the parameters for variant detection.
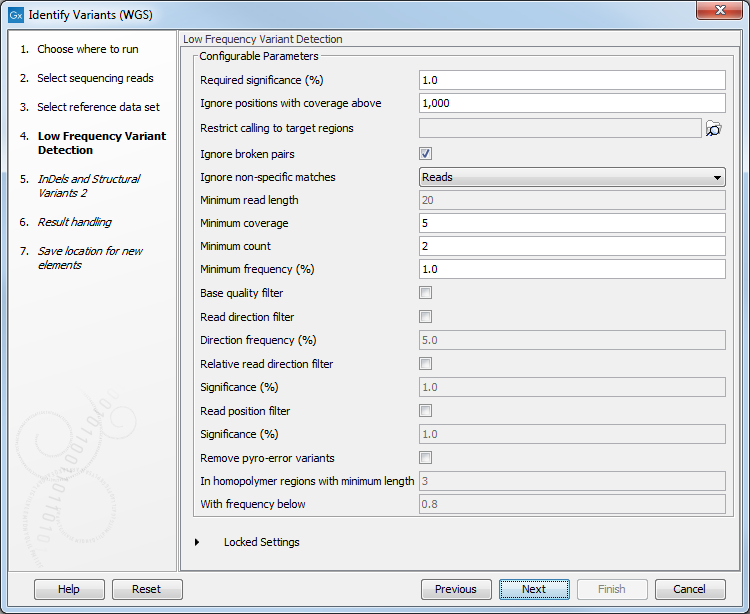
Figure 20.26: Specify the parameters that should be used to detect variants. - In the Indels and Structural Variants 2 dialog (figure 20.27), specify a target regions file if you wish for the variants found outside the targeted region to be removed from the output.
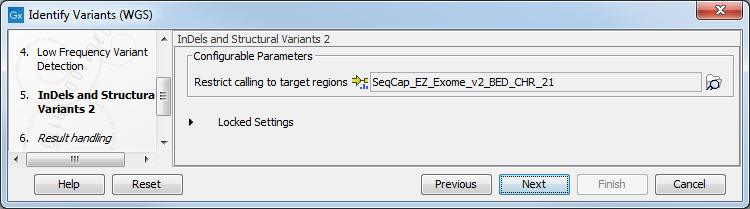
Figure 20.27: Specify the parameters that should be used to detect variants. - In the last wizard step you can check the selected settings by clicking on the button labeled Preview All Parameters.
In the Preview All Parameters wizard you can only check the settings, and if you wish to make changes you have to use the Previous button from the wizard to edit parameters in the relevant windows.
- Choose to Save your results and click Finish.
Output from the Identify Variants (WGS) workflow
The Identify Variants (WGS) tool produces six different types of output:
- Structural Variants (
 ) Variant track showing the structural variants; insertions, deletions, replacements. Hold the mouse over one of the variants or right-clicking on the variant. A tooltip will appear with detailed information about the variant. The structural variants can also be viewed in table format by switching to the table view. This is done by pressing the table icon found in the lower left corner of the View Area.
) Variant track showing the structural variants; insertions, deletions, replacements. Hold the mouse over one of the variants or right-clicking on the variant. A tooltip will appear with detailed information about the variant. The structural variants can also be viewed in table format by switching to the table view. This is done by pressing the table icon found in the lower left corner of the View Area.
- Structural Variant Report (
 ) The report consists of a number of tables and graphs that in different ways provide information about the structural variants.
) The report consists of a number of tables and graphs that in different ways provide information about the structural variants.
- Read Mapping (
 ) The mapped sequencing reads. The reads are shown in different colors depending on their orientation, whether they are single reads or paired reads, and whether they map unambiguously (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Coloring_mapped_reads.html).
) The mapped sequencing reads. The reads are shown in different colors depending on their orientation, whether they are single reads or paired reads, and whether they map unambiguously (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Coloring_mapped_reads.html).
- Read Mapping Report (
 ) The report consists of a number of tables and graphs that in different ways provide information about the mapped reads.
) The report consists of a number of tables and graphs that in different ways provide information about the mapped reads.
- Unfiltered and Filtered Variants (
 ) Variant tracks holding the identified variants before the filters are applied (Unfiltered), and after. Filtered variants are separated into 2 tracks, one for all identified variants, and one containing larger indels. The variants can be shown in track format or in table format. When holding the mouse over the detected variants in the Track List, a tooltip appears with information about the individual variants. You will have to zoom in on the variants to be able to see the detailed tooltip.
) Variant tracks holding the identified variants before the filters are applied (Unfiltered), and after. Filtered variants are separated into 2 tracks, one for all identified variants, and one containing larger indels. The variants can be shown in track format or in table format. When holding the mouse over the detected variants in the Track List, a tooltip appears with information about the individual variants. You will have to zoom in on the variants to be able to see the detailed tooltip.
- Track List (
 ) A collection of tracks presented together. Shows the annotated variant track together with the human reference sequence, genes, transcripts, coding regions, the mapped reads, the identified variants, and the structural variants (see figure 20.5).
) A collection of tracks presented together. Shows the annotated variant track together with the human reference sequence, genes, transcripts, coding regions, the mapped reads, the identified variants, and the structural variants (see figure 20.5).
Before looking at the identified variants, we recommend that you first take a look at the mapping report to see whether the coverage is sufficient in the regions of interest (e.g. > 30 ). Furthermore, please check that at least 90% of the reads map to the human reference sequence. In case of a targeted experiment, please also check that the majority of reads map to the targeted region.
Next, open the Track List (see figure 20.28). It lists the track of the identified variants in context to the human reference sequence, genes, transcripts, coding regions, and mapped sequencing reads.
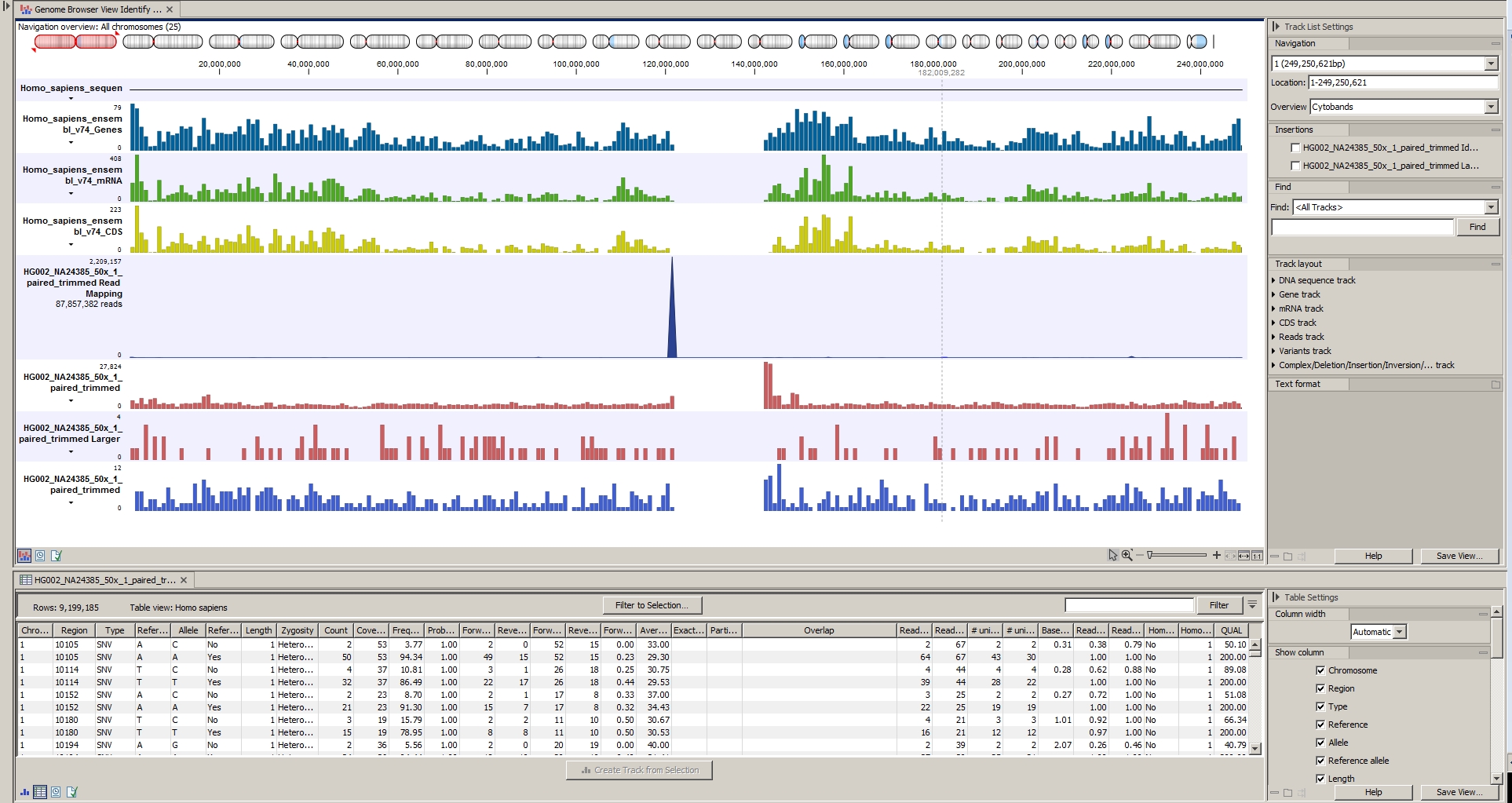
Figure 20.28: The Track List allows easy inspection of the identified smaller variants, larger insertions and deletions, and structural variants in the
context of the human genome.
By double-clicking on the indels variant track in the Track List, a table will be shown that lists all identified larger insertions and deletions (see figure 20.29).
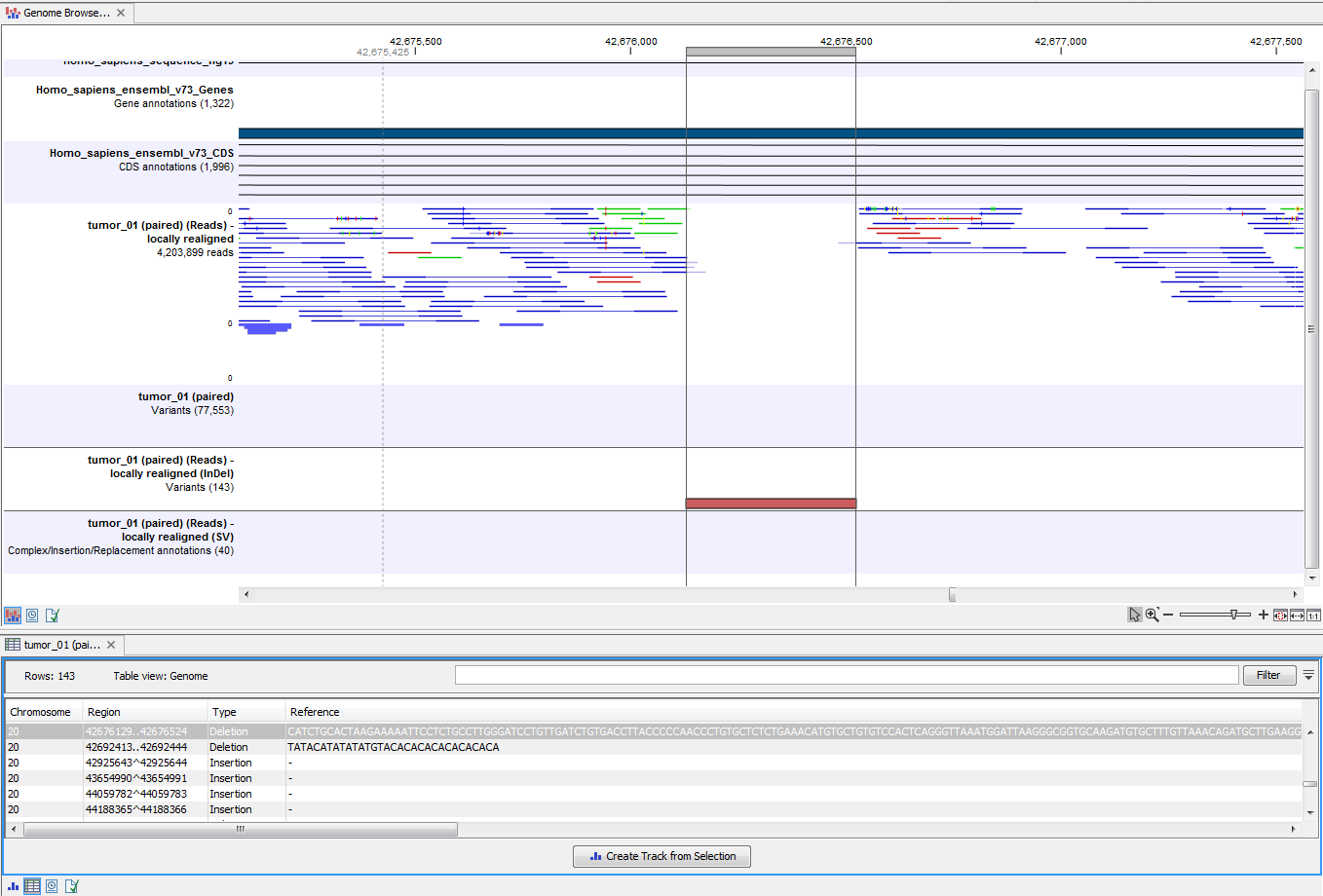
Figure 20.29: This figure shows a Track List with an open track table. The table allows deeper inspection of the identified variants.
