Data import and export
Most import and export of data is done using the tools delivered as part of the CLC Genomics Workbench. This functionality is described at https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Import_export_data_graphics.html.
This chapter describes only import/export functionality specific to the Biomedical Genomics Analysis.
Data import
Data import is generally done using the standard tools of the CLC Genomics Workbench. This includes the on-the-fly import functionality available when launching workflows, described at:
Direct access to certain NGS importers is also provided via the Analyze QIAseq Panels tool, as described in this section.
Reference data import is described in Reference Data Management.
Data export
General data export documentation is available at https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Data_export.html.VCF (Biomedical) exporter
The Biomedical Genomics Analysis plugin provides a VCF exporter that extends the functionality of the standard VCF exporter in the CLC Genomics Workbench by supporting the export of CNV tracks and fusion tracks, in addition to the export of variant tracks.
Compared with the standard VCF exporter, an additional option is present in the export dialog allowing the export of multiple inputs as a single file (figure 2.1). When this option is selected, a CNV track, fusion track and variant tracks can be exported together into a single VCF file.
Where the same variant is reported multiple times, which is especially relevant when providing multiple variant tracks as input, the VCF file will include only one of these, the copy with the highest QUAL value.
Note: When working with fusion data, only fusions with "PASS" in the "Filter" column will be exported.
The standard VCF exporter is described at https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Export_in_VCF_format.html.
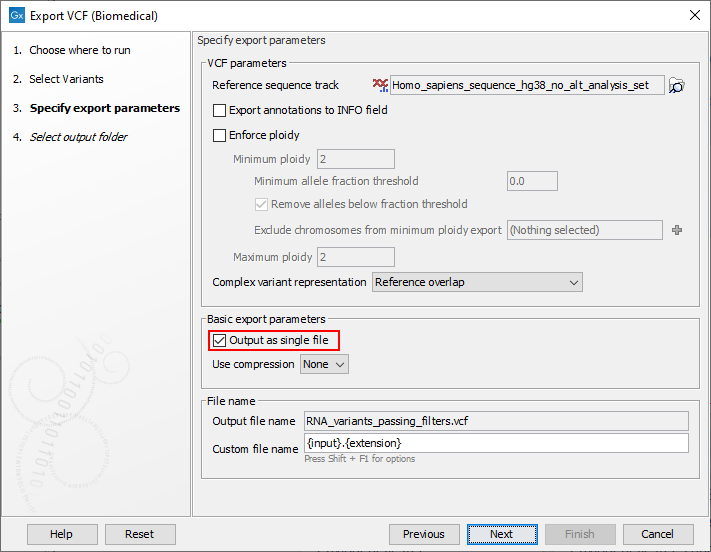
Figure 2.1: The VCF (Biomedical) exporter dialog. The option in red is specific to this exporter. The other options in the dialog are the same as in the standard VCF exporter.
VCF format files can be uploaded directly to QCI Interpret, or the Upload to QCII tool can be used to upload data from CLC Genomics Workbench without explicitly exporting it first. Use of Upload to QCII is described in QCI Interpret Integration.
SAM/BAM export of UMI read mappings When exporting UMI reads to SAM or BAM format, a UMI will be described as Unique_Molecular_Index=[number1]_count=[number2]. Here number1 is a UMI ID (just a unique UMI group number), and number2 is the number of reads that are in that UMI group.
This information is relevant to the export of relevant outputs from workflows distributed with this plugin for analyzing data generated using some types of QIAseq panels, such as:
- Targeted DNA Panels
- Targeted RNAscan Panels
- UPX 3' RNA Panels
- Multimodal Panels
- RNA Fusion XP
